A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila
- PMID: 14871946
- PMCID: PMC329501
- DOI: 10.1128/EC.3.1.157-169.2004
A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila
Abstract
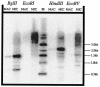
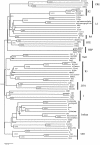
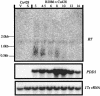
The ciliated protozoan Tetrahymena thermophila undergoes extensive programmed DNA rearrangements during the development of a somatic macronucleus from the germ line micronucleus in its sexual cycle. To investigate the relationship between programmed DNA rearrangements and transposable elements, we identified several members of a family of non-long terminal repeat (LTR) retrotransposons (retroposons) in T. thermophila, the first characterized in the ciliated protozoa. This multiple-copy retrotransposon family is restricted to the micronucleus of T. thermophila. The REP (Tetrahymena non-LTR retroposon) elements encode an ORF2 typical of non-LTR elements that contains apurinic/apyrimidinic endonuclease (APE) and reverse transcriptase (RT) domains. Phylogenetic analysis of the RT and APE domains indicates that the element forms a deep-branching clade within the non-LTR retrotransposon family. Northern analysis with a probe to the conserved RT domain indicates that transcripts from the element are small and heterogeneous in length during early macronuclear development. The presence of a repeated transposable element in the genome is consistent with the model that programmed DNA deletion in T. thermophila evolved as a method of eliminating deleterious transposons from the somatic macronucleus.
Figures





Similar articles
-
Role of micronucleus-limited DNA in programmed deletion of mse2.9 during macronuclear development of Tetrahymena thermophila.Eukaryot Cell. 2004 Apr;3(2):288-301. doi: 10.1128/EC.3.2.288-301.2004. Eukaryot Cell. 2004. PMID: 15075259 Free PMC article.
-
A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila.Nucleic Acids Res. 2002 Jun 1;30(11):2524-37. doi: 10.1093/nar/30.11.2524. Nucleic Acids Res. 2002. PMID: 12034842 Free PMC article.
-
Reverse transcriptase activity and untranslated region sharing of a new RTE-like, non-long terminal repeat retrotransposon from the human blood fluke, Schistosoma japonicum.Int J Parasitol. 2002 Aug;32(9):1163-74. doi: 10.1016/s0020-7519(02)00063-2. Int J Parasitol. 2002. PMID: 12117499
-
DIRS-1 and the other tyrosine recombinase retrotransposons.Cytogenet Genome Res. 2005;110(1-4):575-88. doi: 10.1159/000084991. Cytogenet Genome Res. 2005. PMID: 16093711 Review.
-
Keeping the soma free of transposons: programmed DNA elimination in ciliates.J Biol Chem. 2011 Oct 28;286(43):37045-52. doi: 10.1074/jbc.R111.276964. Epub 2011 Sep 13. J Biol Chem. 2011. PMID: 21914793 Free PMC article. Review.
Cited by
-
The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development.Cell. 2014 Aug 28;158(5):1187-1198. doi: 10.1016/j.cell.2014.07.034. Cell. 2014. PMID: 25171416 Free PMC article.
-
DNA rearrangements directed by non-coding RNAs in ciliates.Wiley Interdiscip Rev RNA. 2010 Nov-Dec;1(3):376-87. doi: 10.1002/wrna.34. Wiley Interdiscip Rev RNA. 2010. PMID: 21956937 Free PMC article. Review.
-
Small-RNA-Mediated Genome-wide trans-Recognition Network in Tetrahymena DNA Elimination.Mol Cell. 2015 Jul 16;59(2):229-42. doi: 10.1016/j.molcel.2015.05.024. Epub 2015 Jun 18. Mol Cell. 2015. PMID: 26095658 Free PMC article.
-
Role of micronucleus-limited DNA in programmed deletion of mse2.9 during macronuclear development of Tetrahymena thermophila.Eukaryot Cell. 2004 Apr;3(2):288-301. doi: 10.1128/EC.3.2.288-301.2004. Eukaryot Cell. 2004. PMID: 15075259 Free PMC article.
-
Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome.Elife. 2016 Nov 28;5:e19090. doi: 10.7554/eLife.19090. Elife. 2016. PMID: 27892853 Free PMC article.
References
-
- Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45-S55. - PubMed
-
- Bibillo, A., and T. H. Eickbush. 2002. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J. Biol. Chem. 277:34836-34845. - PubMed
-
- Borst, P. 2002. Antigenic variation and allelic exclusion. Cell 109:5-8. - PubMed
-
- Borst, P., and D. R. Greaves. 1987. Programmed gene rearrangements altering gene expression. Science 235:658-667. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

