The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases
- PMID: 14960713
- PMCID: PMC384351
- DOI: 10.1093/nar/gkh252
The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases
Abstract
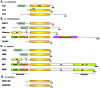
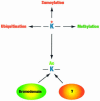
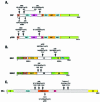
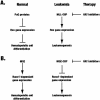
Acetylation of the epsilon-amino group of lysine residues, or N(epsilon)-lysine acetylation, is an important post-translational modification known to occur in histones, transcription factors and other proteins. Since 1995, dozens of proteins have been discovered to possess intrinsic lysine acetyltransferase activity. Although most of these enzymes were first identified as histone acetyltransferases and then tested for activities towards other proteins, acetyltransferases only modifying non-histone proteins have also been identified. Lysine acetyltransferases form different groups, three of which are Gcn5/PCAF, p300/CBP and MYST proteins. While members of the former two groups mainly function as transcriptional co-activators, emerging evidence suggests that MYST proteins, such as Esa1, Sas2, MOF, TIP60, MOZ and MORF, have diverse roles in various nuclear processes. Aberrant lysine acetylation has been implicated in oncogenesis. The genes for p300, CBP, MOZ and MORF are rearranged in recurrent leukemia-associated chromosomal abnormalities. Consistent with their roles in leukemogenesis, these acetyltransferases interact with Runx1 (or AML1), one of the most frequent targets of chromosomal translocations in leukemia. Therefore, the diverse superfamily of lysine acetyltransferases executes an acetylation program that is important for different cellular processes and perturbation of such a program may cause the development of cancer and other diseases.
Figures


References
-
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. - PubMed
-
- Berger S.L. (2001) An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene, 20, 3007–3013. - PubMed
-
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. - PubMed
-
- Turner B.M. (2002) Cellular memory and the histone code. Cell, 111, 285–291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

