Structure-function correlations derived from faster variants of a RNA ligase deoxyribozyme
- PMID: 14960718
- PMCID: PMC373398
- DOI: 10.1093/nar/gkh263
Structure-function correlations derived from faster variants of a RNA ligase deoxyribozyme
Abstract
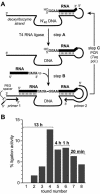
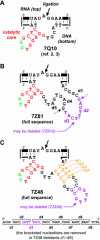
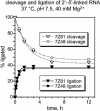
We previously reported the in vitro selection of several Mg2+-dependent deoxyribozymes (DNA enzymes) that synthesize a 2'-5' RNA linkage from a 2',3'-cyclic phosphate and a 5'-hydroxyl. Here we subjected the 9A2 deoxyribozyme to re-selection for improved ligation rate. We found two new DNA enzymes (7Z81 and 7Z48) that contain the catalytic core of 7Q10, a previously reported small deoxyribozyme that is unrelated in sequence to 9A2. A third new DNA enzyme (7Z101) is unrelated to either 7Q10 or 9A2. The new 7Z81 and 7Z48 DNA enzymes have ligation rates over an order of magnitude higher than that of 7Q10 itself and they have additional sequence elements that correlate with these faster rates. Our findings provide insight into structure-function relationships of catalytic nucleic acids.
Figures


References
-
- Flynn-Charlebois A., Wang,Y., Prior,T.K., Rashid,I., Hoadley,K.A., Coppins,R.L., Wolf,A.C. and Silverman,S.K. (2003) Deoxyribozymes with 2′-5′ RNA ligase activity. J. Am. Chem. Soc., 125, 2444–2454. - PubMed
-
- Flynn-Charlebois A., Prior,T.K., Hoadley,K.A. and Silverman,S.K. (2003) In vitro evolution of an RNA-cleaving DNA enzyme into an RNA ligase switches the selectivity from 3′-5′ to 2′-5′. J. Am. Chem. Soc., 125, 5346–5350. - PubMed
-
- Ricca B.L., Wolf,A.C. and Silverman,S.K. (2003) Optimization and generality of a small deoxyribozyme that ligates RNA. J. Mol. Biol., 330, 1015–1025. - PubMed
-
- Wang Y. and Silverman,S.K. (2003) Deoxyribozymes that synthesize branched and lariat RNA. J. Am. Chem. Soc., 125, 6880–6881. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

