Role for CCR5Delta32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells
- PMID: 14963124
- PMCID: PMC369216
- DOI: 10.1128/jvi.78.5.2277-2287.2004
Role for CCR5Delta32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells
Abstract
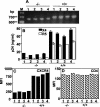
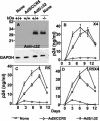
CCR5Delta32 is a loss-of-function mutation that abolishes cell surface expression of the human immunodeficiency virus (HIV) coreceptor CCR5 and provides genetic resistance to HIV infection and disease progression. Since CXCR4 and other HIV coreceptors also exist, we hypothesized that CCR5Delta32-mediated resistance may be due not only to the loss of CCR5 function but also to a gain-of-function mechanism, specifically the active inhibition of alternative coreceptors by the mutant CCR5Delta32 protein. Here we demonstrate that efficient expression of the CCR5Delta32 protein in primary CD4(+) cells by use of a recombinant adenovirus (Ad5/Delta32) was able to down-regulate surface expression of both wild-type CCR5 and CXCR4 and to confer broad resistance to R5, R5X4, and X4 HIV type 1 (HIV-1). This may be important clinically, since we found that CD4(+) cells purified from peripheral blood mononuclear cells of individuals who were homozygous for CCR5Delta32, which expressed the mutant protein endogenously, consistently expressed lower levels of CXCR4 and showed less susceptibility to X4 HIV-1 isolates than cells from individuals lacking the mutation. Moreover, CD4(+) cells from individuals who were homozygous for CCR5Delta32 expressed the mutant protein in five of five HIV-exposed, uninfected donors tested but not in either of two HIV-infected donors tested. The mechanism of inhibition may involve direct scavenging, since we were able to observe a direct interaction of CCR5 and CXCR4 with CCR5Delta32, both by genetic criteria using the yeast two-hybrid system and by biochemical criteria using the coimmunoprecipitation of heterodimers. Thus, these results suggest that at least two distinct mechanisms may account for genetic resistance to HIV conferred by CCR5Delta32: the loss of wild-type CCR5 surface expression and the generation of CCR5Delta32 protein, which functions as a scavenger of both CCR5 and CXCR4.
Figures




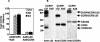
References
-
- Agrawal, L., and G. Alkhatib. 2001. Chemokine receptors: emerging opportunities for new anti-HIV therapies. Expert Opin. Ther. Targets 5:303-326. - PubMed
-
- Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. - PubMed
-
- Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology 234:340-348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

