Absence of replication-competent human-tropic porcine endogenous retroviruses in the germ line DNA of inbred miniature Swine
- PMID: 14963152
- PMCID: PMC369242
- DOI: 10.1128/jvi.78.5.2502-2509.2004
Absence of replication-competent human-tropic porcine endogenous retroviruses in the germ line DNA of inbred miniature Swine
Abstract
The potential transmission of porcine endogenous retroviruses (PERVs) has raised concern in the development of porcine xenotransplantation products. Our previous studies have resulted in the identification of animals within a research herd of inbred miniature swine that lack the capacity to transmit PERV to human cells in vitro. In contrast, other animals were capable of PERV transmission. The PERVs that were transmitted to human cells are recombinants between PERV-A and PERV-C in the post-VRA region of the envelope (B. A. Oldmixon, J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience, J. Virol. 76:3045-3048, 2002); these viruses we term PERV-A/C. This observation prompted us to determine whether these human-tropic replication-competent (HTRC) PERV-A/C recombinants were present in the genomic DNA of these miniature swine. Genomic DNA libraries were generated from one miniature swine that transmitted HTRC PERV as well as from one miniature swine that did not transmit HTRC PERV. HTRC PERV-A/C proviruses were not identified in the germ line DNAs of these pigs by using genomic mapping. Similarly, although PERV-A loci were identified in both libraries that possessed long env open reading frames, the Env proteins encoded by these loci were nonfunctional according to pseudotype assays. In the absence of a germ line source for HTRC PERV, further studies are warranted to assess the mechanisms by which HTRC PERV can be generated. Once identified, it may prove possible to generate animals with further reduced potential to produce HTRC PERV.
Figures


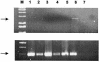
References
-
- Bartosch, B., R. A. Weiss, and Y. Takeuchi. 2002. PCR-based cloning and immunocytological titration of infectious porcine endogenous retrovirus subgroup A and B. J. Gen. Virol. 83:2231-2240. - PubMed
-
- Clark, D. A., J. F. Fryer, A. W. Tucker, P. D. McArdle, A. E. Hughes, V. C. Emery, and P. D. Griffiths. 2003. Porcine cytomegalovirus in pigs being bred for xenograft organs: progress towards control. Xenotransplantation 10:142-148. - PubMed
-
- Dinsmore, J. H., C. Manhart, R. Raineri, D. B. Jacoby, and A. Moore. 2000. No evidence for infection of human cells with porcine endogenous retrovirus (PERV) after exposure to porcine fetal neuronal cells. Transplantation 70:1382-1389. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

