Assembly properties of the human immunodeficiency virus type 1 CA protein
- PMID: 14963157
- PMCID: PMC369201
- DOI: 10.1128/jvi.78.5.2545-2552.2004
Assembly properties of the human immunodeficiency virus type 1 CA protein
Abstract
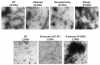
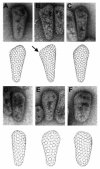
During retroviral maturation, the CA protein oligomerizes to form a closed capsid that surrounds the viral genome. We have previously identified a series of deleterious surface mutations within human immunodeficiency virus type 1 (HIV-1) CA that alter infectivity, replication, and assembly in vivo. For this study, 27 recombinant CA proteins harboring 34 different mutations were tested for the ability to assemble into helical cylinders in vitro. These cylinders are composed of CA hexamers and are structural models for the mature viral capsid. Mutations that diminished CA assembly clustered within helices 1 and 2 in the N-terminal domain of CA and within the crystallographically defined dimer interface in the CA C-terminal domain. These mutations demonstrate the importance of these regions for CA cylinder production and, by analogy, mature capsid assembly. One CA mutant (R18A) assembled into cylinders, cones, and spheres. We suggest that these capsid shapes occur because the R18A mutation alters the frequency at which pentamers are incorporated into the hexagonal lattice. The fact that a single CA protein can simultaneously form all three known retroviral capsid morphologies supports the idea that these structures are organized on similar lattices and differ only in the distribution of 12 pentamers that allow them to close. In further support of this model, we demonstrate that the considerable morphological variation seen for conical HIV-1 capsids can be recapitulated in idealized capsid models by altering the distribution of pentamers.
Figures


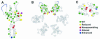
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

