Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation
- PMID: 14963486
- PMCID: PMC380989
- DOI: 10.1038/sj.emboj.7600064
Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation
Abstract
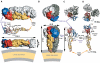
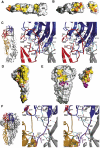
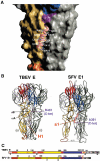
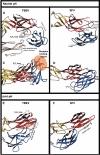
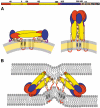
Enveloped viruses enter cells via a membrane fusion reaction driven by conformational changes of specific viral envelope proteins. We report here the structure of the ectodomain of the tick-borne encephalitis virus envelope glycoprotein, E, a prototypical class II fusion protein, in its trimeric low-pH-induced conformation. We show that, in the conformational transition, the three domains of the neutral-pH form are maintained but their relative orientation is altered. Similar to the postfusion class I proteins, the subunits rearrange such that the fusion peptide loops cluster at one end of an elongated molecule and the C-terminal segments, connecting to the viral transmembrane region, run along the sides of the trimer pointing toward the fusion peptide loops. Comparison with the low-pH-induced form of the alphavirus class II fusion protein reveals striking differences at the end of the molecule bearing the fusion peptides, suggesting an important conformational effect of the missing membrane connecting segment.
Figures

References
-
- Bricogne G (1993) Buster. Acta Crystallogr D 49: 37–60 - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: A new software system for macromolecular structure determination. Acta Crystallogr D 54: 905–921 - PubMed
-
- Carson M (1987) Ribbon models of macromolecules. J Mol Graphics 5: 103–106
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

