Psoralen interstrand cross-link repair is specifically altered by an adjacent triple-stranded structure
- PMID: 14966263
- PMCID: PMC373402
- DOI: 10.1093/nar/gkh267
Psoralen interstrand cross-link repair is specifically altered by an adjacent triple-stranded structure
Abstract
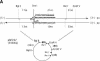
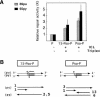
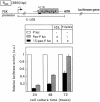
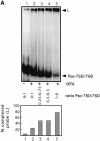
Targeting DNA-damaging agents to specific DNA sites by using sequence-specific DNA ligands has been successful in directing genomic modifications. The understanding of repair processing of such targeted damage and the influence of the adjacent complex is largely unknown. In this way, directed interstrand cross-links (ICLs) have already been generated by psoralen targeting. The mechanisms responsible for ICL removal are far from being understood in mammalian cells, with the proposed involvement of both mutagenic and recombinogenic pathways. Here, a unique ICL was introduced at a selected site by photoactivation of a psoralen moiety with the use of psoralen conjugates of triplex-forming oligonucleotides. The processing of psoralen ICL was evaluated in vitro and in cells for two types of cross-linked substrates, either containing a psoralen ICL alone or with an adjacent triple-stranded structure. We show that the presence of a neighbouring triplex structure interferes with different stages of psoralen ICL processing: (i) the ICL-induced DNA repair synthesis in HeLa cell extracts is inhibited by the triplex structure, as measured by the efficiency of 'true' and futile repair synthesis, stopping at the ICL site; (ii) in HeLa cells, the ICL removal via a nucleotide excision repair (NER) pathway is delayed in the presence of a neighbouring triplex; and (iii) the binding to ICL of recombinant xeroderma pigmentosum A protein, which is involved in pre-incision recruitment of NER factors is impaired by the presence of the third DNA strand. These data characterize triplex-induced modulation of ICL repair pathways at specific steps, which might have implications for the controlled induction of targeted genomic modifications and for the associated cellular responses.
Figures