Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins
- PMID: 14966264
- PMCID: PMC373410
- DOI: 10.1093/nar/gkh278
Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins
Abstract
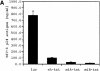
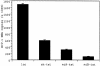
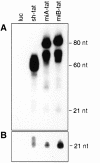
Post-transcriptional inhibition of HIV-1 replication can be achieved by RNA interference (RNAi). The cellular expression of short interfering RNA (siRNA) or short hairpin RNA (shRNA) homologous to regions of the HIV-1 genome decreases viral replication by the selective degradation of targeted RNA. Here, we demonstrate that another class of noncoding regulatory RNA, termed microRNA (miRNA), can be used to deliver antiviral RNAi. By incorporating sequences encoding siRNA targeting the HIV-1 transactivator protein tat into a human miR-30 pre-microRNA (pre-miRNA) backbone, we were able to express tat siRNA in cells. The tat siRNA delivered as pre-miRNA precursor was 80% more effective in reducing HIV-1 p24 antigen production than tat siRNA expressed as conventional shRNA. Our results confirm the utility of expressing HIV-1 specific siRNA through a miR-30 precursor stem-loop structure and suggest that this strategy can be used to increase the antiviral potency of RNAi.
Figures


References
-
- Tuschl T. (2002) Expanding small RNA interference. Nat. Biotechnol., 20, 446–448. - PubMed
-
- Zamore P.D. (2001) RNA interference: listening to the sound of silence. Nature Struct. Biol., 8, 746–750. - PubMed
-
- Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. - PubMed
-
- Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

