Replication protein A (RPA) phosphorylation prevents RPA association with replication centers
- PMID: 14966274
- PMCID: PMC350552
- DOI: 10.1128/MCB.24.5.1930-1943.2004
Replication protein A (RPA) phosphorylation prevents RPA association with replication centers
Abstract
Mammalian replication protein A (RPA) undergoes DNA damage-dependent phosphorylation at numerous sites on the N terminus of the RPA2 subunit. To understand the functional significance of RPA phosphorylation, we expressed RPA2 variants in which the phosphorylation sites were converted to aspartate (RPA2(D)) or alanine (RPA2(A)). Although RPA2(D) was incorporated into RPA heterotrimers and supported simian virus 40 DNA replication in vitro, the RPA2(D) mutant was selectively unable to associate with replication centers in vivo. In cells containing greatly reduced levels of endogenous RPA2, RPA2(D) again did not localize to replication sites, indicating that the defect in supporting chromosomal DNA replication is not due to competition with the wild-type protein. Use of phosphospecific antibodies demonstrated that endogenous hyperphosphorylated RPA behaves similarly to RPA2(D). In contrast, under DNA damage or replication stress conditions, RPA2(D), like RPA2(A) and wild-type RPA2, was competent to associate with DNA damage foci as determined by colocalization with gamma-H2AX. We conclude that RPA2 phosphorylation prevents RPA association with replication centers in vivo and potentially serves as a marker for sites of DNA damage.
Figures

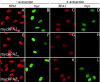

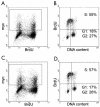





Similar articles
-
Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair.J Biol Chem. 2007 Dec 7;282(49):35910-23. doi: 10.1074/jbc.M704645200. Epub 2007 Oct 10. J Biol Chem. 2007. PMID: 17928296
-
Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes.EMBO J. 1999 Mar 1;18(5):1397-406. doi: 10.1093/emboj/18.5.1397. EMBO J. 1999. PMID: 10064605 Free PMC article.
-
Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress.J Cell Sci. 2009 Nov 15;122(Pt 22):4070-80. doi: 10.1242/jcs.053702. Epub 2009 Oct 20. J Cell Sci. 2009. PMID: 19843584 Free PMC article.
-
Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability.Semin Cell Dev Biol. 2019 Feb;86:112-120. doi: 10.1016/j.semcdb.2018.04.005. Epub 2018 Nov 13. Semin Cell Dev Biol. 2019. PMID: 29665433 Review.
-
Replication protein A phosphorylation and the cellular response to DNA damage.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1015-24. doi: 10.1016/j.dnarep.2004.03.028. DNA Repair (Amst). 2004. PMID: 15279788 Review.
Cited by
-
Role of RPA Phosphorylation in the ATR-Dependent G2 Cell Cycle Checkpoint.Genes (Basel). 2023 Dec 13;14(12):2205. doi: 10.3390/genes14122205. Genes (Basel). 2023. PMID: 38137027 Free PMC article.
-
Dynamic elements of replication protein A at the crossroads of DNA replication, recombination, and repair.Crit Rev Biochem Mol Biol. 2020 Oct;55(5):482-507. doi: 10.1080/10409238.2020.1813070. Epub 2020 Aug 28. Crit Rev Biochem Mol Biol. 2020. PMID: 32856505 Free PMC article. Review.
-
HERC2 regulates RPA2 by mediating ATR-induced Ser33 phosphorylation and ubiquitin-dependent degradation.Sci Rep. 2019 Oct 3;9(1):14257. doi: 10.1038/s41598-019-50812-x. Sci Rep. 2019. PMID: 31582797 Free PMC article.
-
RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response.J Biol Chem. 2011 Jun 24;286(25):22314-22. doi: 10.1074/jbc.M111.222802. Epub 2011 May 9. J Biol Chem. 2011. PMID: 21558276 Free PMC article.
-
Mitotic crisis: the unmasking of a novel role for RPA.Cell Cycle. 2009 Feb 1;8(3):357-61. doi: 10.4161/cc.8.3.7496. Epub 2009 Feb 21. Cell Cycle. 2009. PMID: 19176996 Free PMC article. Review.
References
-
- Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. - PubMed
-
- Bartek, J., and J. Lukas. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13:738-747. - PubMed
-
- Binz, S. K., Y. Lao, D. F. Lowry, and M. S. Wold. 2003. The phosphorylation domain of the 32-kDa subunit of replication protein A modulates RPA-DNA interactions: evidence for an intersubunit interaction. J. Biol. Chem. 278:35584-35591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
