Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1
- PMID: 14966286
- PMCID: PMC350571
- DOI: 10.1128/MCB.24.5.2074-2082.2004
Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1
Abstract
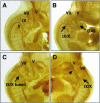
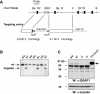
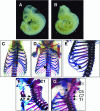
LMO4 belongs to a family of transcriptional regulators that comprises two zinc-binding LIM domains. LIM-only (LMO) proteins appear to function as docking sites for other factors, leading to the assembly of multiprotein complexes. The transcription factor Deaf-1/NUDR has been identified as one partner protein of LMO4. We have disrupted the Lmo4 and Deaf-1 genes in mice to define their biological function in vivo. All Lmo4 mutants died shortly after birth and showed defects within the presphenoid bone, with 50% of mice also exhibiting exencephaly. Homeotic transformations were observed in Lmo4-null embryos and newborn mice, but with incomplete penetrance. These included skeletal defects in cervical vertebrae and the rib cage. Furthermore, fusions of cranial nerves IX and X and defects in cranial nerve V were apparent in some Lmo4(-/-) and Lmo4(+/-) mice. Remarkably, Deaf-1 mutants displayed phenotypic abnormalities similar to those observed in Lmo4 mutants. These included exencephaly, transformation of cervical segments, and rib cage abnormalities. In contrast to Lmo4 nullizygous mice, nonexencephalic Deaf-1 mutants remained healthy. No defects in the sphenoid bone or cranial nerves were apparent. Thus, Lmo4 and Deaf-1 mutant mice exhibit overlapping as well as distinct phenotypes. Our data indicate an important role for these two transcriptional regulators in pathways affecting neural tube closure and skeletal patterning, most likely reflecting their presence in a functional complex in vivo.
Figures




References
-
- Agulnick, A. D., M. Taira, J. J. Breen, T. Tanaka, I. B. Dawid, and H. Westphal. 1996. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384:270-272. - PubMed
-
- Bach, I. 2000. The LIM domain: regulation by association. Mech. Dev. 91:5-17. - PubMed
-
- Bach, I., C. Carriere, H. P. Ostendorff, B. Andersen, and M. G. Rosenfeld. 1997. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11:1370-1380. - PubMed
-
- Bottomley, M. J., M. W. Collard, J. I. Huggenvik, Z. Liu, T. J. Gibson, and M. Sattler. 2001. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8:626-633. - PubMed
-
- Chai, Y., X. Jiang, Y. Ito, P. Bringas, Jr., J. Han, D. H. Rowitch, P. Soriano, A. P. McMahon, and H. M. Sucov. 2000. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127:1671-1679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
