Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead
- PMID: 14966295
- PMCID: PMC350549
- DOI: 10.1128/MCB.24.5.2181-2189.2004
Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead
Abstract
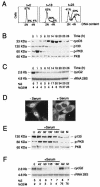
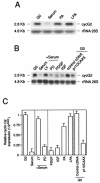
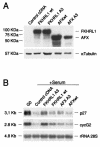
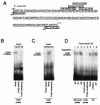
Cyclin G2 is an unconventional cyclin highly expressed in postmitotic cells. Unlike classical cyclins that promote cell cycle progression, cyclin G2 blocks cell cycle entry. Here we studied the mechanisms that regulate cyclin G2 mRNA expression during the cell cycle. Analysis of synchronized NIH 3T3 cell cultures showed elevated cyclin G2 mRNA expression levels at G(0), with a considerable reduction as cells enter cell cycle. Downregulation of cyclin G2 mRNA levels requires activation of phosphoinositide 3-kinase, suggesting that this enzyme controls cyclin G2 mRNA expression. Because the phosphoinositide 3-kinase pathway inhibits the FoxO family of forkhead transcription factors, we examined the involvement of these factors in the regulation of cyclin G2 expression. We show that active forms of the forkhead transcription factor FoxO3a (FKHRL1) increase cyclin G2 mRNA levels. Cyclin G2 has forkhead consensus motifs in its promoter, which are transactivated by constitutive active FoxO3a forms. Finally, interference with forkhead-mediated transcription by overexpression of an inactive form decreases cyclin G2 mRNA expression levels. These results show that FoxO genes regulate cyclin G2 expression, illustrating a new role for phosphoinositide 3-kinase and FoxO transcription factors in the control of cell cycle entry.
Figures


References
-
- Alvarez, B., E. Garrido, J. A. Garcia-Sanz, and A. C. Carrera. 2003. Phosphoinositide 3-kinase activation regulates cell division time by coordinated control of cell mass and cell cycle progression rate. J. Biol. Chem. 27:26466-26473. - PubMed
-
- Alvarez, B., C. Martinez-A., B. M. T. Burgering, and A. C. Carrera. 2001. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413:744-747. - PubMed
-
- Bennin, D. A., A. S. Don, T. Brake, J. L. McKenzie, H. Rosenbaum, L. Ortiz, A. A. DePaoli-Roach, and M. C. Horne. 2002. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B′ subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J. Biol. Chem. 277:27449-27467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
