Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome
- PMID: 14966531
- PMCID: PMC340940
- DOI: 10.1371/journal.pbio.0020029
Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome
Abstract
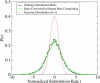
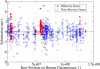
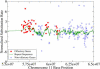
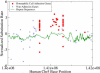
The neutral mutation rate is known to vary widely along human chromosomes, leading to mutational hot and cold regions. We provide evidence that categories of functionally related genes reside preferentially in mutationally hot or cold regions, the size of which we have measured. Genes in hot regions are biased toward extracellular communication (surface receptors, cell adhesion, immune response, etc.), while those in cold regions are biased toward essential cellular processes (gene regulation, RNA processing, protein modification, etc.). From a selective perspective, this organization of genes could minimize the mutational load on genes that need to be conserved and allow fast evolution for genes that must frequently adapt. We also analyze the effect of gene duplication and chromosomal recombination, which contribute significantly to these biases for certain categories of hot genes. Overall, our results show that genes are located nonrandomly with respect to hot and cold regions, offering the possibility that selection acts at the level of gene location in the human genome.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures

References
-
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, et al. Vol. 1. New York: Garland Publishing; 1994. Molecular biology of the cell; 616 pp.
-
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. - PubMed
-
- Casane D, Boissinot S, Chang BH-J, Shimmin LC, Li WH. Mutation pattern among regions of the primate genome. J Mol Evol. 1997;45:216–226. - PubMed

