The yeast coexpression network has a small-world, scale-free architecture and can be explained by a simple model
- PMID: 14968131
- PMCID: PMC1299002
- DOI: 10.1038/sj.embor.7400090
The yeast coexpression network has a small-world, scale-free architecture and can be explained by a simple model
Abstract
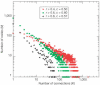
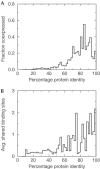
We investigated the gene coexpression network in Saccharomyces cerevisiae, in which genes are linked when they are coregulated. This network is shown to have a scale-free, small-world architecture. Such architecture is typical of biological networks in which the nodes are connected when they are involved in the same biological process. Current models for the evolution of intracellular networks do not adequately reproduce the features that we observe in the network. We therefore derive a new model for its evolution based on the observation that there is a positive correlation between the sequence similarity of paralogues and their probability of coexpression or sharing of transcription factor binding sites (TFBSs). The simple, neutralist's model consists of (1) coduplication of genes with their TFBSs, (2) deletion and duplication of individual TFBSs and (3) gene loss. A network is constructed by connecting genes that share multiple TFBSs. Our model reproduces the scale-free, small-world architecture of the coregulation network and the homology relations between coregulated genes without the need for selection either at the level of the network structure or at the level of gene regulation.
Figures


References
-
- Aloy P, Russell RB ( 2002) Potential artefacts in protein-interaction networks. FEBS Lett 530: 253–254 - PubMed
-
- Barabasi AL, Albert R ( 1999) Emergence of scaling in random networks. Science 286: 509–512 - PubMed
-
- Bhan A, Galas DJ, Dewey TG ( 2002) A duplication growth model of gene expression networks. Bioinformatics 18: 1486–1493 - PubMed
-
- Brummelkamp TR, Bernards R ( 2003) New tools for functional mammalian cancer genetics. Nat Rev Cancer 3: 781–789 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

