Convergent evolution of gene networks by single-gene duplications in higher eukaryotes
- PMID: 14968135
- PMCID: PMC1299007
- DOI: 10.1038/sj.embor.7400096
Convergent evolution of gene networks by single-gene duplications in higher eukaryotes
Abstract
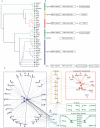
By combining phylogenetic, proteomic and structural information, we have elucidated the evolutionary driving forces for the gene-regulatory interaction networks of basic helix-loop-helix transcription factors. We infer that recurrent events of single-gene duplication and domain rearrangement repeatedly gave rise to distinct networks with almost identical hub-based topologies, and multiple activators and repressors. We thus provide the first empirical evidence for scale-free protein networks emerging through single-gene duplications, the dominant importance of molecular modularity in the bottom-up construction of complex biological entities, and the convergent evolution of networks.
Figures

References
-
- Apic G, Gough J, Teichmann SA ( 2001) Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol 310: 311–325 - PubMed
-
- Barabasi A ( 2002) Linked, the New Science of Networks. Oxford: Perseus Press
-
- Beckmann H, Kadesch T ( 1991) The leucine zipper of TFE3 dictates helix–loop–helix dimerization specificity. Genes Dev 5: 1057–1066 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

