Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA
- PMID: 14970383
- PMCID: PMC1370933
- DOI: 10.1261/rna.5169404
Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA
Abstract
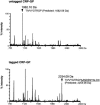
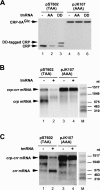
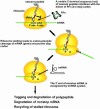
Recent studies have established that tmRNA-mediated protein tagging occurs at stop codons depending on the C-terminal amino acid sequence of the nascent polypeptide immediately adjacent to those codons. We investigate here how the trans-translation at a stop codon occurs by using model crp genes encoding variants of cAMP receptor protein (CRP). We demonstrate that a truncated crp mRNA is efficiently produced along with a normal transcript from the model gene where tmRNA-mediated protein tagging occurs. The truncated crp mRNA was not detected in the presence of tmRNA, indicating that its degradation was facilitated by tmRNA. The major 3'-ends of the truncated crp mRNA in cells unable to express tmRNA were mapped at and near the stop codon. When RNA derived from the model crp-crr fusion gene was analyzed, crr mRNA was detected as a downstream cleavage product along with the upstream crp mRNA. These results are compatible with the hypothesis that ribosome stalling caused by the tagging-provoking sequences leads to endonucleolytic cleavage of mRNA around the stop codon, resulting in nonstop mRNA. In addition, the data are consistent with the view that mRNA cleavage is the cause of trans-translation at stop codons. Neither the bacterial toxin RelE nor the known major endoribonucleases are required for this cleavage, indicating that either other endoribonuclease(s) or the ribosome itself would be responsible for the mRNA cleavage in response to ribosome stalling caused by the particular nascent peptides.
Figures



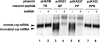


References
-
- Abe, H., Abo, T., and Aiba, H. 1999. Regulation of intrinsic terminator by translation in Escherichia coli: Transcription termination at a distance downstream. Genes Cells 4: 87–97. - PubMed
-
- Abo, T., Ueda, K., Sunohara, T., Ogawa, K., and Aiba, H. 2002. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells 7: 629–638. - PubMed
-
- Aiba, H., Adhya, S., and de Crombrugghe, B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256: 11905–11910. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
