Interaction of the Bacillus subtilis RNase P with the 30S ribosomal subunit
- PMID: 14970393
- PMCID: PMC1370943
- DOI: 10.1261/rna.5163104
Interaction of the Bacillus subtilis RNase P with the 30S ribosomal subunit
Abstract
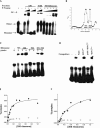
Ribonuclease P (RNase P) is a ribozyme required for the 5' maturation of all tRNA. RNase P and the ribosome are the only known ribozymes conserved in all organisms. We set out to determine whether this ribonucleoprotein enzyme interacts with other cellular components, which may imply other functions for this conserved ribozyme. Incubation of the Bacillus subtilis RNase P holoenzyme with fractionated B. subtilis cellular extracts and purified ribosomal subunits results in the formation of a gel-shifted complex with the 30S ribosomal subunit at a binding affinity of approximately 40 nM in 0.1 M NH(4)Cl and 10 mM MgCl(2). The complex does not form with the RNase P RNA alone and is disrupted by a mRNA mimic polyuridine, but is stable in the presence of high concentrations of mature tRNA. Endogenous RNase P can also be detected in the 30S ribosomal fraction. Cleavage of a pre-tRNA substrate by the RNase P holoenzyme remains the same in the presence of the 30S ribosome, but the cleavage of an artificial non-tRNA substrate is inhibited eightfold. Hydroxyl radical protection and chemical modification identify several protected residues located in a highly conserved region in the RNase P RNA. A single mutation within this region significantly reduces binding, providing strong support on the specificity of the RNase P-30S ribosome complex. Our results also suggest that the dimeric form of the RNase P is primarily involved in 30S ribosome binding. We discuss several models on a potential function of the RNase P-30S ribosome complex.
Figures
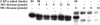


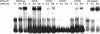
References
-
- Alifano, P., Rivellini, F., Piscitelli, C., Arraiano, C.M., Bruni, C.B., and Carlomagno, M.S. 1994. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes & Dev. 8: 3021–3031. - PubMed
-
- Altman, S. and Kirsebom, L. 1999. Ribonuclease P. In The RNA world, 2nd ed. (eds. R.F. Gesteland et al.), pp. 351–380. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Barrera, A., Fang, X., Jacob, J., Casey, E., Thiyagarajan, P., Pan, T. 2002. Dimeric and monomeric Bacillus subtilis RNase P holoenzyme in the absence and presence of pre-tRNA substrates. Biochemistry 41: 12986–12994. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
