Development of surface adhesion in Caulobacter crescentus
- PMID: 14973013
- PMCID: PMC344395
- DOI: 10.1128/JB.186.5.1438-1447.2004
Development of surface adhesion in Caulobacter crescentus
Abstract
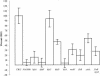
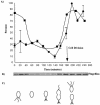
Caulobacter crescentus has a dimorphic life cycle composed of a motile stage and a sessile stage. In the sessile stage, C. crescentus is often found tightly attached to a surface through its adhesive holdfast. In this study, we examined the contribution of growth and external structures to the attachment of C. crescentus to abiotic surfaces. We show that the holdfast is essential but not sufficient for optimal attachment. Rather, adhesion in C. crescentus is a complex developmental process. We found that the attachment of C. crescentus to surfaces is cell cycle regulated and that growth or energy or both are essential for this process. The initial stage of attachment occurs in swarmer cells and is facilitated by flagellar motility and pili. Our results suggest that strong attachment is mediated by the synthesis of a holdfast as the swarmer cell differentiates into a stalked cell.
Figures


References
-
- Brun, Y., G. Marczynski, and L. Shapiro. 1994. The expression of asymmetry during cell differentiation. Annu. Rev. Biochem. 63:419-450. - PubMed
-
- Brun, Y. V., and L. J. Shimkets. 2000. Prokaryotic development. American Society for Microbiology, Washington, D.C.
-
- Cole, J., G. G. Hardy, D. Bodenmiller, E. Toh, A. Hinz, and Y. V. Brun. 2003. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol. Microbiol. 49:1671-1683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

