Assembly of an oxalate decarboxylase produced under sigmaK control into the Bacillus subtilis spore coat
- PMID: 14973022
- PMCID: PMC344410
- DOI: 10.1128/JB.186.5.1462-1474.2004
Assembly of an oxalate decarboxylase produced under sigmaK control into the Bacillus subtilis spore coat
Abstract
Over 30 polypeptides are synthesized at various times during sporulation in Bacillus subtilis, and they are assembled at the surface of the developing spore to form a multilayer protein structure called the coat. The coat consists of three main layers, an amorphous undercoat close to the underlying spore cortex peptidoglycan, a lamellar inner layer, and an electron-dense striated outer layer. The product of the B. subtilis oxdD gene was previously shown to have oxalate decarboxylase activity when it was produced in Escherichia coli and to be a spore constituent. In this study, we found that OxdD specifically associates with the spore coat structure, and in this paper we describe regulation of its synthesis and assembly. We found that transcription of oxdD is induced during sporulation as a monocistronic unit under the control of sigma(K) and is negatively regulated by GerE. We also found that localization of a functional OxdD-green fluorescent protein (GFP) at the surface of the developing spore depends on the SafA morphogenetic protein, which localizes at the interface between the spore cortex and coat layers. OxdD-GFP localizes around the developing spore in a cotE mutant, which does not assemble the spore outer coat layer, but it does not persist in spores produced by the mutant. Together, the data suggest that OxdD-GFP is targeted to the interior layers of the coat. Additionally, we found that expression of a multicopy allele of oxdD resulted in production of spores with increased levels of OxdD that were able to degrade oxalate but were sensitive to lysozyme.
Figures

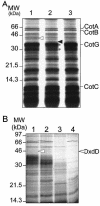
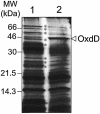




References
-
- Anand, R., P. C. Dorrestein, C. Kinsland, T. P. Begley, and S. E. Ealick. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution. Biochemistry 41:7659-7669. - PubMed
-
- Aronson, A. I., L. Ekanayake, and P. C. Fitz-James. 1992. Protein filaments may initiate the assembly of the Bacillus subtilis spore coat. Biochimie 74:661-667. - PubMed
-
- Aronson, A. I., H. Y. Song, and N. Bourne. 1989. Gene structure and precursor processing of a novel Bacillus subtilis spore coat protein. Mol. Microbiol. 3:437-444. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

