Chitinase B of "Microbulbifer degradans" 2-40 contains two catalytic domains with different chitinolytic activities
- PMID: 14973034
- PMCID: PMC344425
- DOI: 10.1128/JB.186.5.1297-1303.2004
Chitinase B of "Microbulbifer degradans" 2-40 contains two catalytic domains with different chitinolytic activities
Abstract
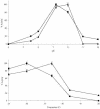
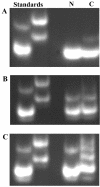
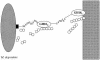
Chitinase B of "Microbulbifer degradans" 2-40 is a modular protein that is predicted to contain two glycoside hydrolase family 18 (GH18) catalytic domains, two polyserine domains, and an acidic repeat domain. Each of the GH18 domains was shown to be catalytically active against chitin. Activity assays reveal that the amino-terminal catalytic domain (GH18(N)) releases methylumbelliferone from 4'-methylumbelliferyl-N,N'-diacetylchitobiose 13.6-fold faster than the carboxy-terminal catalytic domain (GH18(C)) and releases chitobiose from the nonreducing end of chitooligosaccharides, therefore functioning as an exochitinase. GH18(C) releases methylumbelliferone from 4'-methylumbelliferyl-N,N',N"-triacetylchitotriose 2.7-fold faster than GH18(N) and cleaves chitooligosaccharides at multiple bonds, consistent with endochitinolytic activity. Each domain was maximally active from 30 to 37 degrees C and from pH 7.2 to 8.0 and was not affected by Mg(2+), Mn(2+), Ca(2+), K(+), EDTA, EGTA, or 1.0 M NaCl. The activity of each domain was moderately inhibited by Ni(2+), Sr(2+), and Cu(2+), while Hg(2+) completely abolished activity. When the specific activities of various recombinant portions of ChiB were calculated by using native chitin as a substrate, the polypeptide containing the endo-acting domain was twofold more active on native chitin than the other containing the exo-acting domain. The presence of both domains in a single reaction increased the amount of reducing sugars released from native chitin to 140% above the theoretical combined rate, indicating that the domains function cooperatively to degrade chitin. These data demonstrate that the GH18 domains of ChiB have different activities on the same substrate and function cooperatively to enhance chitin depolymerization.
Figures


Similar articles
-
The deduced role of a chitinase containing two nonsynergistic catalytic domains.Acta Crystallogr D Struct Biol. 2018 Jan 1;74(Pt 1):30-40. doi: 10.1107/S2059798317018289. Epub 2018 Jan 1. Acta Crystallogr D Struct Biol. 2018. PMID: 29372897 Free PMC article.
-
Characterization of two novel bacterial type A exo-chitobiose hydrolases having C-terminal 5/12-type carbohydrate-binding modules.Appl Microbiol Biotechnol. 2017 Jun;101(11):4533-4546. doi: 10.1007/s00253-017-8198-4. Epub 2017 Mar 9. Appl Microbiol Biotechnol. 2017. PMID: 28280871
-
Genomic analysis and initial characterization of the chitinolytic system of Microbulbifer degradans strain 2-40.J Bacteriol. 2003 Jun;185(11):3352-60. doi: 10.1128/JB.185.11.3352-3360.2003. J Bacteriol. 2003. PMID: 12754233 Free PMC article.
-
Chitinases: biomarkers for human diseases.Protein Pept Lett. 2009;16(5):490-8. doi: 10.2174/092986609788167842. Protein Pept Lett. 2009. PMID: 19442228 Review.
-
The broad-specificity chitinases: their origin, characterization, and potential application.Appl Microbiol Biotechnol. 2019 Apr;103(8):3289-3295. doi: 10.1007/s00253-019-09718-x. Epub 2019 Mar 8. Appl Microbiol Biotechnol. 2019. PMID: 30850873 Review.
Cited by
-
Identification and characterization of human polyserase-3, a novel protein with tandem serine-protease domains in the same polypeptide chain.BMC Biochem. 2006 Mar 27;7:9. doi: 10.1186/1471-2091-7-9. BMC Biochem. 2006. PMID: 16566820 Free PMC article.
-
Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40 T.PLoS Genet. 2008 May 30;4(5):e1000087. doi: 10.1371/journal.pgen.1000087. PLoS Genet. 2008. PMID: 18516288 Free PMC article.
-
Chitinase gene sequences retrieved from diverse aquatic habitats reveal environment-specific distributions.Appl Environ Microbiol. 2004 Dec;70(12):6977-83. doi: 10.1128/AEM.70.12.6977-6983.2004. Appl Environ Microbiol. 2004. PMID: 15574890 Free PMC article.
-
Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production.Bioengineering (Basel). 2017 Jun 11;4(2):55. doi: 10.3390/bioengineering4020055. Bioengineering (Basel). 2017. PMID: 28952534 Free PMC article. Review.
-
Polymer degrading marine Microbulbifer bacteria: an un(der)utilized source of chemical and biocatalytic novelty.Beilstein J Org Chem. 2024 Jul 17;20:1635-1651. doi: 10.3762/bjoc.20.146. eCollection 2024. Beilstein J Org Chem. 2024. PMID: 39076296 Free PMC article. Review.
References
-
- Brurberg, M. B., I. F. Nes, and V. G. Eijsink. 1996. Comparative studies of chitinases A and B from Serratia marcescens. Microbiology 142:1581-1589. - PubMed
-
- Ensor, L., S. Stosz, and R. Weiner. 1999. Expression of multiple complex polysaccharide-degrading enzyme systems by marine bacterium strain 2-40. J. Ind. Microbiol. Biotechnol. 23:123-126. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

