The switch I and II regions of MinD are required for binding and activating MinC
- PMID: 14973039
- PMCID: PMC344430
- DOI: 10.1128/JB.186.5.1546-1555.2004
The switch I and II regions of MinD are required for binding and activating MinC
Abstract
MinD and MinC cooperate to form an efficient inhibitor of Z-ring formation that is spatially regulated by MinE. MinD activates MinC by recruiting it to the membrane and targeting it to a septal component. To better understand this activation, we have isolated loss-of-function mutations in minD and carried out site-directed mutagenesis. Many of these mutations block MinC-MinD interaction; however, they also prevent MinD self-interaction and membrane binding, suggesting that they affect nucleotide interaction or protein folding. Two mutations in the switch I region (MinD box) and one mutation in the switch II region had little affect on most MinD functions, such as MinD self-interaction, membrane binding, and MinE stimulation; however, they did eliminate MinD-MinC interaction. Two additional mutations in the switch II region did not affect MinC binding. Further study revealed that one of these allowed the MinCD complex to target to the septum but was still deficient in blocking division. These results indicate that the switch I and II regions of MinD are required for interaction with MinC but not MinE and that the switch II region has a role in activating MinC.
Figures

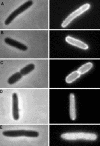
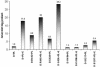


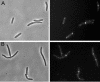
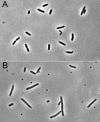

Similar articles
-
Analysis of MinD mutations reveals residues required for MinE stimulation of the MinD ATPase and residues required for MinC interaction.J Bacteriol. 2005 Jan;187(2):629-38. doi: 10.1128/JB.187.2.629-638.2005. J Bacteriol. 2005. PMID: 15629934 Free PMC article.
-
Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE.J Bacteriol. 2003 Jan;185(1):196-203. doi: 10.1128/JB.185.1.196-203.2003. J Bacteriol. 2003. PMID: 12486056 Free PMC article.
-
A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum.Mol Microbiol. 2003 Jan;47(2):345-55. doi: 10.1046/j.1365-2958.2003.03321.x. Mol Microbiol. 2003. PMID: 12519187
-
MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation.Mol Microbiol. 2003 Apr;48(2):295-303. doi: 10.1046/j.1365-2958.2003.03427.x. Mol Microbiol. 2003. PMID: 12675792 Review.
-
Spatial control of the cell division site by the Min system in Escherichia coli.Environ Microbiol. 2013 Dec;15(12):3229-39. doi: 10.1111/1462-2920.12119. Epub 2013 Apr 9. Environ Microbiol. 2013. PMID: 23574354 Review.
Cited by
-
A new multicompartmental reaction-diffusion modeling method links transient membrane attachment of E. coli MinE to E-ring formation.Syst Synth Biol. 2010 Mar;4(1):35-53. doi: 10.1007/s11693-009-9047-2. Epub 2009 Dec 10. Syst Synth Biol. 2010. PMID: 20012222 Free PMC article.
-
Emerging facets of plastid division regulation.Planta. 2013 Feb;237(2):389-98. doi: 10.1007/s00425-012-1743-6. Epub 2012 Sep 11. Planta. 2013. PMID: 22965912 Review.
-
Plastid division: evolution, mechanism and complexity.Ann Bot. 2007 Apr;99(4):565-79. doi: 10.1093/aob/mcl249. Epub 2006 Nov 30. Ann Bot. 2007. PMID: 17138581 Free PMC article. Review.
-
Functional conservation of the MIN plastid division homologues of Chlamydomonas reinhardtii.Planta. 2008 May;227(6):1199-211. doi: 10.1007/s00425-008-0692-6. Epub 2008 Feb 13. Planta. 2008. PMID: 18270733
-
Role of MinD-membrane association in Min protein interactions.J Bacteriol. 2006 Apr;188(8):2993-3001. doi: 10.1128/JB.188.8.2993-3001.2006. J Bacteriol. 2006. PMID: 16585760 Free PMC article.
References
-
- Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. - PubMed
-
- Cordell, S. C., and J. Lowe. 2001. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 492:160-165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

