Transcriptional organization and regulation of the L-idonic acid pathway (GntII system) in Escherichia coli
- PMID: 14973046
- PMCID: PMC344402
- DOI: 10.1128/JB.186.5.1388-1397.2004
Transcriptional organization and regulation of the L-idonic acid pathway (GntII system) in Escherichia coli
Abstract
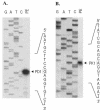
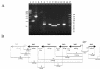
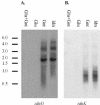
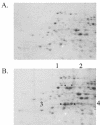
The genetic organization of the idn genes that encode the pathway for L-idonate catabolism was characterized. The monocistronic idnK gene is transcribed divergently from the idnDOTR genes, which were shown to form an operon. The 215-bp regulatory region between the idnK and idnD genes contains promoters in opposite orientation with transcription start sites that mapped to positions -26 and -29 with respect to the start codons. The regulatory region also contains a single putative IdnR/GntR binding site centered between the two promoters, a CRP binding site upstream of idnD, and an UP element upstream of idnK. The genes of the L-idonate pathway were shown to be under catabolite repression control. Analysis of idnD- and idnK-lacZ fusions in a nonpolar idnD mutant that is unable to interconvert L-idonate and 5-ketogluconate indicated that either compound could induce the pathway. The L-idonate pathway was first characterized as a subsidiary pathway for D-gluconate catabolism (GntII), which is induced by D-gluconate in a GntI (primary gluconate system) mutant. Here we showed that the idnK and idnD operons are induced by D-gluconate in a GntI system mutant, presumably by endogenous formation of 5-ketogluconate from D-gluconate. Thus, the regulation of the GntII system is appropriate for this pathway, which is primarily involved in L-idonate catabolism; the GntII system can be induced by D-gluconate under conditions that block the GntI system.
Figures

Similar articles
-
Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli.J Bacteriol. 1998 Jul;180(14):3704-10. doi: 10.1128/JB.180.14.3704-3710.1998. J Bacteriol. 1998. PMID: 9658018 Free PMC article.
-
The activator of GntII genes for gluconate metabolism, GntH, exerts negative control of GntR-regulated GntI genes in Escherichia coli.J Bacteriol. 2003 Mar;185(6):1783-95. doi: 10.1128/JB.185.6.1783-1795.2003. J Bacteriol. 2003. PMID: 12618441 Free PMC article.
-
Dual control by regulators, GntH and GntR, of the GntII genes for gluconate metabolism in Escherichia coli.J Mol Microbiol Biotechnol. 2003;6(1):41-56. doi: 10.1159/000073407. J Mol Microbiol Biotechnol. 2003. PMID: 14593252
-
The subsidiary GntII system for gluconate metabolism in Escherichia coli: alternative induction of the gntV gene.Biol Res. 2011;44(3):269-75. Epub 2011 Nov 7. Biol Res. 2011. PMID: 22688914
-
D-galactonate metabolism in enteric bacteria: a molecular and physiological perspective.Curr Opin Microbiol. 2024 Oct;81:102524. doi: 10.1016/j.mib.2024.102524. Epub 2024 Aug 12. Curr Opin Microbiol. 2024. PMID: 39137493 Review.
Cited by
-
Deletion of genes involved in the ketogluconate metabolism, Entner-Doudoroff pathway, and glucose dehydrogenase increase local and invasive virulence phenotypes in Streptococcus pneumoniae.PLoS One. 2019 Jan 8;14(1):e0209688. doi: 10.1371/journal.pone.0209688. eCollection 2019. PLoS One. 2019. PMID: 30620734 Free PMC article.
-
Development of a Genome-Scale Metabolic Model and Phenome Analysis of the Probiotic Escherichia coli Strain Nissle 1917.Int J Mol Sci. 2021 Feb 20;22(4):2122. doi: 10.3390/ijms22042122. Int J Mol Sci. 2021. PMID: 33672760 Free PMC article.
-
Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum.J Bacteriol. 2006 Jan;188(2):409-23. doi: 10.1128/JB.188.2.409-423.2006. J Bacteriol. 2006. PMID: 16385030 Free PMC article.
-
Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources.Infect Immun. 2005 Dec;73(12):8039-49. doi: 10.1128/IAI.73.12.8039-8049.2005. Infect Immun. 2005. PMID: 16299298 Free PMC article.
-
Revealing the genetic basis of natural bacterial phenotypic divergence.J Bacteriol. 2014 Feb;196(4):825-39. doi: 10.1128/JB.01039-13. Epub 2013 Dec 6. J Bacteriol. 2014. PMID: 24317396 Free PMC article.
References
-
- Bachi, B., and H. L. Kornberg. 1975. Genes involved in the uptake and catabolism of gluconate by Escherichia coli. J. Gen. Microbiol. 90:321-335. - PubMed
-
- Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Conway, T., B. Kraus, D. L. Tucker, D. J. Smalley, A. F. Dorman, and L. McKibben. 2002. DNA array analysis in a Microsoft Windows environment. BioTechniques 32:110, 112-114, 116, 118-119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

