Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers
- PMID: 14973191
- PMCID: PMC365734
- DOI: 10.1073/pnas.0307323101
Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers
Abstract
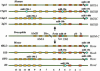
A large number of tiny noncoding RNAs have been cloned and named microRNAs (miRs). Recently, we have reported that miR-15a and miR-16a, located at 13q14, are frequently deleted and/or down-regulated in patients with B cell chronic lymphocytic leukemia, a disorder characterized by increased survival. To further investigate the possible involvement of miRs in human cancers on a genome-wide basis, we have mapped 186 miRs and compared their location to the location of previous reported nonrandom genetic alterations. Here, we show that miR genes are frequently located at fragile sites, as well as in minimal regions of loss of heterozygosity, minimal regions of amplification (minimal amplicons), or common breakpoint regions. Overall, 98 of 186 (52.5%) of miR genes are in cancer-associated genomic regions or in fragile sites. Moreover, by Northern blotting, we have shown that several miRs located in deleted regions have low levels of expression in cancer samples. These data provide a catalog of miR genes that may have roles in cancer and argue that the full complement of miRs in a genome may be extensively involved in cancers.
Figures



References
-
- Ke, X.-S., Liu, C.-M., Liu, D.-P. & Liang, C.-C. (2003) Curr. Opin. Chem. Biol. 7, 516-523. - PubMed
-
- Moss, E. G. (2003) MicroRNAs in Noncoding RNAs: Molecular Biology and Molecular Medicine (Eurekah.com), pp. 98-114.
-
- Pasquinelli, A. E., Reinhart, B. J., Slack F., Martindale, M. Q., Kuroda, M. I., Maller, B., Hayward, D. C., Ball, E. E., Degnan, B., Muller, P., et al. (2000) Nature 408, 86-89. - PubMed
-
- Ambros, V. (2003) Cell 113, 673-676. - PubMed
-
- Xu, P., Vernooy, S. Y., Guo, M. & Hay, B. A. (2003) Curr. Biol. 13, 790-795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

