FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis
- PMID: 14973192
- PMCID: PMC365781
- DOI: 10.1073/pnas.0306778101
FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis
Abstract
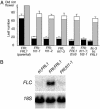
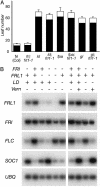
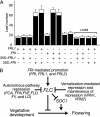
In temperate climates, the prolonged cold temperature of winter serves as a seasonal landmark for winter-annual and biennial plants. In these plants, flowering is blocked before winter. In Arabidopsis thaliana, natural variation in the FRIGIDA (FRI) gene is a major determinate of the rapid-cycling vs. winter-annual flowering habits. In winter-annual accessions of Arabidopsis, FRI activity blocks flowering through the up-regulation of the floral inhibitor FLOWERING LOCUS C (FLC). Most rapid-flowering accessions, in contrast, contain null alleles of FRI. By performing a mutant screen in a winter-annual strain, we have identified a locus, FRIGIDA LIKE 1 (FRL1), that is specifically required for the up-regulation of FLC by FRI. Cloning of FRL1 revealed a gene with a predicted protein sequence that is 23% identical to FRI. Despite sequence similarity, FRI and FRL1 do not have redundant functions. FRI and FRL1 belong to a seven-member gene family in Arabidopsis, and FRI, FRL1, and at least one additional family member, FRIGIDA LIKE 2 (FRL2), are in a clade of this family that is required for the winter-annual habit in Arabidopsis.
Figures

References
-
- Corre, V. L., Roux, F. & Reboud, X. (2002) Mol. Biol. Evol. 19, 1261-1271. - PubMed
-
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R. & Dean, C. (2000) Science 290, 344-347. - PubMed
-
- Napp-Zinn, K. (1979) in La Physiologie de la Floraison, eds. Champagnat, P. & Jaques, R. (Collogues Internationaux Centre National de la Recherche Scientifique, Paris), pp. 217-220.
-
- Lee, I., Bleecker, A. & Amasino, R. (1993) Mol. Gen. Genet. 237, 171-176. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

