Portability and fidelity of RNA-repair systems
- PMID: 14973195
- PMCID: PMC365698
- DOI: 10.1073/pnas.0305859101
Portability and fidelity of RNA-repair systems
Abstract
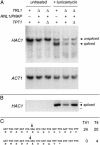
Yeast tRNA ligase (Trl1) is an essential enzyme that converts cleaved tRNA half-molecules into spliced tRNAs containing a 2'-PO(4), 3'-5' phosphodiester at the splice junction. Trl1 also catalyzes splicing of HAC1 mRNA during the unfolded protein response. Trl1 performs three reactions: the 2',3'-cyclic phosphate of the proximal RNA fragment is hydrolyzed to a 3'-OH, 2'-PO(4) by a cyclic phosphodiesterase; the 5'-OH of the distal RNA fragment is phosphorylated by a GTP-dependent polynucleotide kinase; and the 3'-OH, 2'-PO(4), and 5'-PO(4) ends are then sealed by an ATP-dependent RNA ligase. The removal of the 2'-PO(4) at the splice junction is catalyzed by the essential enzyme Tpt1, which transfers the RNA 2'-PO(4) to NAD(+) to form ADP-ribose 1"-2"-cyclic phosphate. Here, we show that the bacteriophage T4 enzymes RNA ligase 1 and polynucleotide kinase/phosphatase can fulfill the tRNA and HAC1 mRNA splicing functions of yeast Trl1 in vivo and bypass the requirement for Tpt1. These results attest to the portability of RNA-repair systems, notwithstanding the significant differences in the specificities, mechanisms, and reaction intermediates of the individual yeast and T4 enzymes responsible for the RNA healing and sealing steps. We surmise that Tpt1 and its unique metabolite ADP-ribose 1"-2"-cyclic phosphate do not play essential roles in yeast independent of the tRNA-splicing reaction. Our finding that one-sixth of spliced HAC1 mRNAs in yeast cells containing the T4 RNA-repair system suffered deletion of a single nucleotide at the 3' end of the splice-donor site suggests a model whereby the yeast RNA-repair system evolved a requirement for the 2'-PO(4) for RNA ligation to suppress inappropriate RNA recombination.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

