Identification of NH...N hydrogen bonds by magic angle spinning solid state NMR in a double-stranded RNA associated with myotonic dystrophy
- PMID: 14973225
- PMCID: PMC373415
- DOI: 10.1093/nar/gkh288
Identification of NH...N hydrogen bonds by magic angle spinning solid state NMR in a double-stranded RNA associated with myotonic dystrophy
Abstract
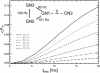
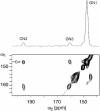
RNA plays a central role in biological processes and exhibits a variety of secondary and tertiary structural features that are often stabilized via hydrogen bonds. The distance between the donor and acceptor nitrogen nuclei involved in NH...N hydrogen bonds in nucleic acid base pairs is typically in the range of 2.6-2.9 A. Here, we show for the first time that such spatial proximity between 15N nitrogen nuclei can be conveniently monitored via magic angle spinning solid state NMR on a uniformly 15N-labelled RNA. The presence of NH.N hydrogen bonds is reflected as cross-peaks between the donor and acceptor nitrogen nuclei in 2D 15N dipolar chemical shift correlation spectra. The RNA selected for this experimental study was a CUG repeat expansion implicated in the neuromuscular disease myotonic dystrophy. The results presented provide direct evidence that the CUG repeat expansion adopts a double-stranded conformation.
Figures




References
-
- Gesteland R.F., Atkins,J.F. and Cech,T.R. (1999) The RNA World (2nd edn). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Ohlenschlager O., Wohnert,J., Ramachandran,R., Sich,C. and Gorlach,M. (2003) Nuclear magnetic resonance studies of ribonucleic acids. Spectroscopy, 17, 537–547.
-
- Furtig B., Richter,C., Wohnert,J. and Schwalbe,H. (2003) NMR spectroscopy of RNA. Chembiochem, 4, 936–962. - PubMed
-
- Ohlenschlager O., Wohnert,J., Bucci,E., Seitz,S., Hafner,S., Ramachandran,R., Zell,R. and Gorlach,M. (2004) The structure of the stemloop D subdomain of Coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure, in press. - PubMed

