Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach
- PMID: 14973327
- PMCID: PMC373429
- DOI: 10.1093/nar/gnh035
Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach
Abstract
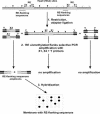
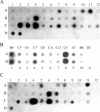
A technique for simultaneous determination of the methylation status of numerous loci containing retroelements (REs) is reported. It is based on the observation that methylated and unmethylated areas in the genome are usually extended, and therefore the methylation of particular methyl-sensitive restriction endonuclease recognition sites might reflect the methylation status of DNA regions around them. The method includes dot-blot hybridization of repeat flanking sequences arrayed on a solid support with specifically amplified flanking regions of presumably unmethylated repeats. A multitude of flanking regions of REs adjacent to unmethylated restriction sites are amplified simultaneously, providing a complex hybridization probe. The technique thus allows the determination of the methylation status of restriction sites, which serve as tags of the methylation status of the surrounding regions. The validity of the technique was confirmed by various means, including bisulfite sequencing. The technique was successfully applied to the identification of methylation patterns of the regions surrounding 38 human-specific HERV-K(HML-2) long terminal repeats in cerebellum- and lymph node-derived genomic DNAs. The described technique can be readily adapted to the use of DNA microarray technology.
Figures

Similar articles
-
Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes.Genetics. 2007 Dec;177(4):1975-85. doi: 10.1534/genetics.107.080234. Genetics. 2007. PMID: 18073417 Free PMC article.
-
A tag-based approach for high-throughput analysis of CCWGG methylation.Anal Biochem. 2007 Oct 15;369(2):154-60. doi: 10.1016/j.ab.2007.06.048. Epub 2007 Jul 5. Anal Biochem. 2007. PMID: 17706584
-
High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum.Nucleic Acids Res. 2009 Jul;37(13):4331-40. doi: 10.1093/nar/gkp393. Epub 2009 May 20. Nucleic Acids Res. 2009. PMID: 19458156 Free PMC article.
-
Methylation of endogenous human retroelements in health and disease.Curr Top Microbiol Immunol. 2006;310:211-50. doi: 10.1007/3-540-31181-5_11. Curr Top Microbiol Immunol. 2006. PMID: 16909913 Review.
-
Techniques used in studies of age-related DNA methylation changes.Ann N Y Acad Sci. 2006 May;1067:479-87. doi: 10.1196/annals.1354.069. Ann N Y Acad Sci. 2006. PMID: 16804030 Review.
Cited by
-
Polymorphism of a new Ty1-copia retrotransposon in durum wheat under salt and light stresses.Theor Appl Genet. 2010 Jul;121(2):311-22. doi: 10.1007/s00122-010-1311-z. Epub 2010 Mar 17. Theor Appl Genet. 2010. PMID: 20237753
-
Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors?Retrovirology. 2013 Feb 9;10:16. doi: 10.1186/1742-4690-10-16. Retrovirology. 2013. PMID: 23394165 Free PMC article. Review.
-
Retroviral Elements in Pathophysiology and as Therapeutic Targets for Amyotrophic Lateral Sclerosis.Neurotherapeutics. 2022 Jul;19(4):1085-1101. doi: 10.1007/s13311-022-01233-8. Epub 2022 Apr 12. Neurotherapeutics. 2022. PMID: 35415778 Free PMC article. Review.
-
Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes.Genetics. 2007 Dec;177(4):1975-85. doi: 10.1534/genetics.107.080234. Genetics. 2007. PMID: 18073417 Free PMC article.
-
Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders.J Virol. 2005 Sep;79(17):10890-901. doi: 10.1128/JVI.79.17.10890-10901.2005. J Virol. 2005. PMID: 16103141 Free PMC article.
References
-
- Jaenisch R. and Bird,A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet., 33 Suppl., 245–254. - PubMed
-
- Bird A.P. (1986) CpG-rich islands and the function of DNA methylation. Nature, 321, 209–213. - PubMed
-
- Turker M.S. (2002) Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene, 21, 5388–5393. - PubMed
-
- vandeLagemaat L.N., Landry,J.R., Mager,D.L. and Medstrand,P. (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet., 19, 530–536. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

