Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreading exonuclease activities: implications for genotyping assays
- PMID: 14973328
- PMCID: PMC373430
- DOI: 10.1093/nar/gnh036
Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreading exonuclease activities: implications for genotyping assays
Abstract
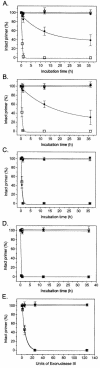
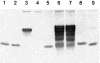
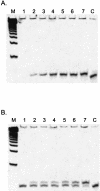
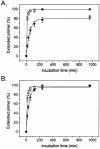
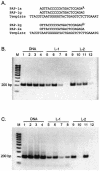
The effect of locked nucleic acid (LNA) modification position upon representative DNA polymerase and exonuclease activities has been examined for potential use in primer extension genotyping applications. For the 3'-->5' exonuclease activities of four proofreading DNA polymerases (Vent, Pfu, Klenow fragment and T7 DNA polymerase) as well as exonuclease III, an LNA at the terminal (L-1) position of a primer is found to provide partial protection against the exonucleases of the two family B polymerases only. In contrast, an LNA residue at the penultimate (L-2) position generates essentially complete nuclease resistance. The polymerase active sites of these enzymes also display a distinct preference. An L-1 LNA modification has modest effects upon poly merization, but an L-2 LNA group slows dTTP incorporation somewhat while virtually abolishing extension with ddTTP or acyTTP terminators, even with A488L Vent DNA polymerase engineered for terminator incorporation. These observations on active site preference have been utilized to demonstrate two novel assays: exonuclease-mediated single base extension (E-SBE) and proofreading allele-specific extension (PRASE). We show that a model PRASE genotyping reaction with L-2 LNA primers offers greater specificity than existing non-proofreading assays, whether or not the non-proofreading reaction employs LNA-modified primers.
Figures

References
-
- Singh S.K., Nielsen,P., Koshkin,A.A. and Wengel,J. (1998) LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun., 4, 455–456.
-
- Obika S., Uneda,T., Sugimoto,T., Nanbu,D., Minami,T., Doi,T. and Imanishi,T. (2001) 2′-O,4′-C-methylene bridged nucleic acid (2′,4′-BNA): synthesis and triplex-forming properties. Bioorg. Med. Chem., 9, 1001–1011. - PubMed
-
- Morita K., Hasegawa,C., Kaneko,M., Tsutsumi,S., Sone,J., Ishikawa,T. and Imanishi,T. (2002) 2′-O,4′-C-ethylene-bridged nucleic acid (ENA): highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drugs. Bioorg. Med. Chem. Lett., 12, 73–76. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

