Unlocking hidden genomic sequence
- PMID: 14973330
- PMCID: PMC373418
- DOI: 10.1093/nar/gnh022
Unlocking hidden genomic sequence
Abstract
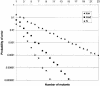
Despite the success of conventional Sanger sequencing, significant regions of many genomes still present major obstacles to sequencing. Here we propose a novel approach with the potential to alleviate a wide range of sequencing difficulties. The technique involves extracting target DNA sequence from variants generated by introduction of random mutations. The introduction of mutations does not destroy original sequence information, but distributes it amongst multiple variants. Some of these variants lack problematic features of the target and are more amenable to conventional sequencing. The technique has been successfully demonstrated with mutation levels up to an average 18% base substitution and has been used to read previously intractable poly(A), AT-rich and GC-rich motifs.
Figures






References
-
- Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. The International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
-
- Glöckner G., Eichinger,L., Szafranski,K., Pachebat,J.A., Bankier,A.T., Dear,P.H., Lehmann,R., Baumgart,C., Parra,G., Abril,J.F. et al. (2002) Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature, 418, 79–85. - PubMed
-
- Donlin M.J. and Johnson,K.A. (1994) Mutants affecting nucleotide recognition by T7 DNA polymerase. Biochemistry, 33, 14908–14917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

