Generating in vitro transcripts with homogenous 3' ends using trans-acting antigenomic delta ribozyme
- PMID: 14973333
- PMCID: PMC373431
- DOI: 10.1093/nar/gnh037
Generating in vitro transcripts with homogenous 3' ends using trans-acting antigenomic delta ribozyme
Abstract
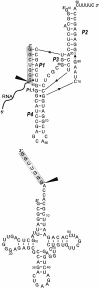
In most in vitro run-off transcription reactions with T7 RNA polymerase, transcripts with heterogeneous ends are commonly obtained. Towards the goal of finding a simple and effective procedure for correct processing of their 3' ends we propose the use of trans-acting antigenomic delta ribozyme. We demonstrate that the extension of nascent transcripts with only seven nucleotides complementary to the ribozyme's recognition site, and subsequently, the removal of those nucleotides with the ribozyme acting in trans, is an efficient procedure for generating transcripts with homogenous 3' ends. This approach was tested on two model RNA molecules: an in vitro transcript of yeast tRNA(Phe) and a delta ribozyme, which processed itself during transcription. The proposed procedure is a simple alternative to the use of ribozymes as cis-cleaving autocatalytic cassettes attached to transcript 3' ends. As there is little possibility that the required additional stretch, only seven nucleotides long, enters into stable interactions with other parts of the transcripts, it can be cleaved off with high efficacy.
Figures


Similar articles
-
Trans-acting antigenomic HDV ribozyme for production of in vitro transcripts with homogenous 3' ends.Methods Mol Biol. 2012;941:99-111. doi: 10.1007/978-1-62703-113-4_8. Methods Mol Biol. 2012. PMID: 23065556
-
cis-Acting 5' hammerhead ribozyme optimization for in vitro transcription of highly structured RNAs.Methods Mol Biol. 2014;1086:21-40. doi: 10.1007/978-1-62703-667-2_2. Methods Mol Biol. 2014. PMID: 24136596
-
Cis-acting ribozymes for the production of RNA in vitro transcripts with defined 5' and 3' ends.Methods Mol Biol. 2012;941:83-98. doi: 10.1007/978-1-62703-113-4_7. Methods Mol Biol. 2012. PMID: 23065555
-
Design and properties of trans-acting HDV ribozymes with extended substrate recognition regions.Nucleic Acids Res Suppl. 2001;(1):201-2. doi: 10.1093/nass/1.1.201. Nucleic Acids Res Suppl. 2001. PMID: 12836334
-
Polyadenylation of rRNA- and tRNA-based yeast transcripts cleaved by internal ribozyme activity.Curr Genet. 2003 Jul;43(4):255-62. doi: 10.1007/s00294-003-0401-8. Epub 2003 May 14. Curr Genet. 2003. PMID: 12748813
Cited by
-
Antigenomic delta ribozyme variants with mutations in the catalytic core obtained by the in vitro selection method.Nucleic Acids Res. 2006 Mar 2;34(4):1270-80. doi: 10.1093/nar/gkl018. Print 2006. Nucleic Acids Res. 2006. PMID: 16513845 Free PMC article.
-
An in-vitro transcription assay for development of Rotavirus VP7.Iran J Microbiol. 2017 Jun;9(3):186-194. Iran J Microbiol. 2017. PMID: 29225758 Free PMC article.
-
A robust and versatile method for production and purification of large-scale RNA samples for structural biology.RNA. 2020 Aug;26(8):1023-1037. doi: 10.1261/rna.075697.120. Epub 2020 Apr 30. RNA. 2020. PMID: 32354720 Free PMC article.
-
An RNA toolbox for single-molecule force spectroscopy studies.Nucleic Acids Res. 2007;35(19):6625-39. doi: 10.1093/nar/gkm585. Epub 2007 Sep 28. Nucleic Acids Res. 2007. PMID: 17905817 Free PMC article.
-
A novel 4-base-recognizing RNA cutter that can remove the single 3' terminal nucleotides from RNA molecules.Nucleic Acids Res. 2004 Jun 28;32(11):e91. doi: 10.1093/nar/gnh092. Print 2004. Nucleic Acids Res. 2004. PMID: 15247324 Free PMC article.
References
-
- Price S.R., Ito,N., Outbridge,C., Avis,J.M. and Nagai,K. (1995) Crystallization of RNA–protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol., 249, 398–408. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

