In vivo interactions of the Acanthamoeba TBP gene promoter
- PMID: 14976219
- PMCID: PMC390285
- DOI: 10.1093/nar/gkh297
In vivo interactions of the Acanthamoeba TBP gene promoter
Abstract
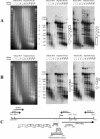
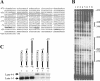
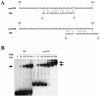
Transcription of the TATA box binding protein (TBP) gene in Acanthamoeba castellanii is regulated by TATA box binding protein promoter binding factor (TPBF), which binds to an upstream TBP promoter element to stimulate transcription, and to a TATA proximal element, where it represses transcription. In order to extend these observations to the in vivo chromatin context, the TBP gene was examined by in situ footprinting and chromatin immunoprecipitation (ChIP). Acanthamoeba DNA is nucleosomal with a repeat of approximately 160 bp, and an intranucleosomal DNA periodicity of 10.5 bp. The TBP gene comprises a 220 bp micrococcal nuclease hypersensitive site corresponding to the promoter regulatory elements previously identified, flanked by protected regions of a size consistent with the presence of nucleosomes. ChIP data indicated that TPBF is associated with the TBP, TPBF and MIL gene promoters, but not to the CSP21, MIIHC, 5SrRNA or 39SrRNA promoters, or to the MIL gene C-terminal region. Binding by TPBF to the TPBF and MIL gene promoters was confirmed by in vitro assays. These results validate the in vitro model for TBP gene regulation and further suggest that TPBF may be autoregulated and may participate in the regulation of the MIL gene.
Figures


Similar articles
-
Purification and characterization of TATA-binding protein promoter binding factor. A regulatory transcription factor of the tbp gene.J Biol Chem. 1994 Jul 15;269(28):18541-8. J Biol Chem. 1994. PMID: 8034602
-
An upstream promoter element of the Acanthamoeba castellanii TBP gene binds a DNA sequence specific transcription activating protein, TPBF.Nucleic Acids Res. 1993 Sep 11;21(18):4321-9. doi: 10.1093/nar/21.18.4321. Nucleic Acids Res. 1993. PMID: 8414988 Free PMC article.
-
Transcription of the Acanthamoeba TATA-binding protein gene. A single transcription factor acts both as an activator and a repressor.J Biol Chem. 1997 Feb 7;272(6):3852-9. doi: 10.1074/jbc.272.6.3852. J Biol Chem. 1997. PMID: 9013645
-
Cloning, expression, and characterization of the TATA-binding protein (TBP) promoter binding factor, a transcription activator of the Acanthamoeba TBP gene.J Biol Chem. 1995 Dec 1;270(48):28839-47. doi: 10.1074/jbc.270.48.28839. J Biol Chem. 1995. PMID: 7499409
-
Recombinant and native Plasmodium falciparum TATA-binding-protein binds to a specific TATA box element in promoter regions.Mol Biochem Parasitol. 2005 Apr;140(2):183-96. doi: 10.1016/j.molbiopara.2005.01.002. Mol Biochem Parasitol. 2005. PMID: 15760658
Cited by
-
Target acquired: transcriptional regulators as drug targets for protozoan parasites.Int J Parasitol. 2021 Jul;51(8):599-611. doi: 10.1016/j.ijpara.2020.12.007. Epub 2021 Mar 13. Int J Parasitol. 2021. PMID: 33722681 Free PMC article. Review.
-
Expression plasmids and production of EGFP in stably transfected Acanthamoeba.Protein Expr Purif. 2010 Mar;70(1):95-100. doi: 10.1016/j.pep.2009.10.008. Epub 2009 Oct 28. Protein Expr Purif. 2010. PMID: 19836453 Free PMC article.
-
N-myc downstream-regulated gene 2, a novel estrogen-targeted gene, is involved in the regulation of Na+/K+-ATPase.J Biol Chem. 2011 Sep 16;286(37):32289-99. doi: 10.1074/jbc.M111.247825. Epub 2011 Jul 19. J Biol Chem. 2011. PMID: 21771789 Free PMC article.
References
-
- Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. - PubMed
-
- Struhl K. (1995) Yeast transcriptional regulatory mechanisms. Annu. Rev. Genet., 29, 651–674. - PubMed
-
- Zawel L. and Reinberg,D. (1993) Initiation of transcription by RNA polymerase II: a multi-step process. Prog. Nucleic Acid Res. Mol. Biol., 44, 67–108. - PubMed
-
- Zawel L. and Reinberg,D. (1995) Common themes in assembly and function of eukaryotic transcription complexes. Annu. Rev. Biochem., 64, 533–561. - PubMed
-
- Ptashne M. and Gann,A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. - PubMed

