Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula
- PMID: 14976232
- PMCID: PMC389921
- DOI: 10.1104/pp.103.030221
Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula
Abstract
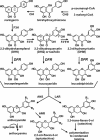
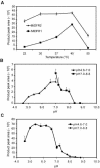
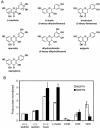
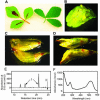
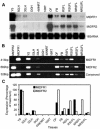
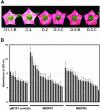
Dihydroflavonol-4-reductase (DFR; EC1.1.1.219) catalyzes a key step late in the biosynthesis of anthocyanins, condensed tannins (proanthocyanidins), and other flavonoids important to plant survival and human nutrition. Two DFR cDNA clones (MtDFR1 and MtDFR2) were isolated from the model legume Medicago truncatula cv Jemalong. Both clones were functionally expressed in Escherichia coli, confirming that both encode active DFR proteins that readily reduce taxifolin (dihydroquercetin) to leucocyanidin. M. truncatula leaf anthocyanins were shown to be cyanidin-glucoside derivatives, and the seed coat proanthocyanidins are known catechin and epicatechin derivatives, all biosynthesized from leucocyanidin. Despite high amino acid similarity (79% identical), the recombinant DFR proteins exhibited differing pH and temperature profiles and differing relative substrate preferences. Although no pelargonidin derivatives were identified in M. truncatula, MtDFR1 readily reduced dihydrokaempferol, consistent with the presence of an asparagine residue at a location known to determine substrate specificity in other DFRs, whereas MtDFR2 contained an aspartate residue at the same site and was only marginally active on dihydrokaempferol. Both recombinant DFR proteins very efficiently reduced 5-deoxydihydroflavonol substrates fustin and dihydrorobinetin, substances not previously reported as constituents of M. truncatula. Transcript accumulation for both genes was highest in young seeds and flowers, consistent with accumulation of condensed tannins and leucoanthocyanidins in these tissues. MtDFR1 transcript levels in developing leaves closely paralleled leaf anthocyanin accumulation. Overexpression of MtDFR1 in transgenic tobacco (Nicotiana tabacum) resulted in visible increases in anthocyanin accumulation in flowers, whereas MtDFR2 did not. The data reveal unexpected properties and differences in two DFR proteins from a single species.
Figures

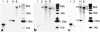


References
-
- Almeida J, Carpenter R, Robbins TP, Martin C, Coen ES (1989) Genetic interactions underlying flower color patterns in Antirrhinun majus. Genes Dev 3: 1758-1767 - PubMed
-
- Baggett BR, Cooper JD, Hogan ET, Carper J, Paiva NL, Smith JT (2002) Profiling isoflavonoids found in legume root extracts using capillary electrophoresis. Electrophoresis 23: 1642-1651 - PubMed
-
- Beld M, Martin C, Huits H, Stuitje AR, Gerats AG (1989) Flavonoid synthesis in Petunia hybrida: partial characterization of dihydroflavonol-4-reductase genes. Plant Mol Biol 13: 491-502 - PubMed
-
- Bongue-Bartelsman M, O'Neill SD, Tong Y, Yoder JI (1994) Characterization of the gene encoding dihydroflavonol 4-reductase in tomato. Gene 138: 153-157 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

