Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis
- PMID: 14976237
- PMCID: PMC389945
- DOI: 10.1104/pp.103.035832
Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis
Abstract
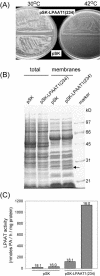
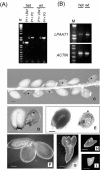
Lysophosphatidyl acyltransferase (LPAAT) is a pivotal enzyme controlling the metabolic flow of lysophosphatidic acid into different phosphatidic acids in diverse tissues. A search of the Arabidopsis genome database revealed five genes that could encode LPAAT-like proteins. We identified one of them, LPAAT1, to be the lone gene that encodes the plastid LPAAT. LPAAT1 could functionally complement a bacterial mutant that has defective LPAAT. Bacteria transformed with LPAAT1 produced LPAAT that had in vitro enzyme activity much higher on 16:0-coenzyme A than on 18:1-coenzyme A in the presence of 18:1-lysophosphatidic acid. LPAAT1 transcript was present in diverse organs, with the highest level in green leaves. A mutant having a T-DNA inserted into LPAAT1 was identified. The heterozygous mutant has no overt phenotype, and its leaf acyl composition is similar to that of the wild type. Selfing of a heterozygous mutant produced normal-sized and shrunken seeds in the Mendelian ratio of 3:1, and the shrunken seeds could not germinate. The shrunken seeds apparently were homozygous of the T-DNA-inserted LPAAT1, and development of the embryo within them was arrested at the heart-torpedo stage. This embryo lethality could be rescued by transformation of the heterozygous mutant with a 35S:LPAAT1 construct. The current findings of embryo death in the homozygous knockout mutant of the plastid LPAAT contrasts with earlier findings of a normal phenotype in the homozygous mutant deficient of the plastid glycerol-3-phosphate acyltransferase; both mutations block the synthesis of plastid phosphatidic acid. Reasons for the discrepancy between the contrasting phenotypes of the two mutants are discussed.
Figures





References
-
- Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen J, Paddock T, Salas J, Savage L, Milcamps A et al. (2003) Arabidopsis thaliana genes involved in acyl lipid metabolism: a 2003 census of the candidates, a study of the distribution of ESTs in organs and a Web-based database. Plant Physiol 132: 681-697 - PMC - PubMed
-
- Bouvier-Nave P, Benveniste P, Oelkers P, Sturley SL, Schaller H (2000) Expression in yeast and tobacco of plant cDNAs encoding acyl CoA: diacylglycerol acyltransferase. Eur J Biochem 267: 85-96 - PubMed
-
- Brown AP, Brough CL, Kroon JTM, Slabas AR (1995) Identification of a cDNA that encodes a 1-acyl-sn-glycerol-3-phosphate acyltransferase from Limnanthes douglasii. Plant Mol Biol 29: 267-278 - PubMed
-
- Brown AP, Coleman J, Tommey AM, Watson MD, Slabas AR (1994) Isolation and characterization of a maize cDNA that complements a 1-acyl-sn-glycerol-3-phosphate acyltaransferase mutant of Escherichia coli and encodes a protein which similarities to other acyltransferases. Plant Mol Biol 26: 211-223 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

