Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses
- PMID: 14976549
- PMCID: PMC381004
- DOI: 10.1038/sj.emboj.7600096
Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses
Abstract
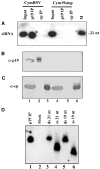
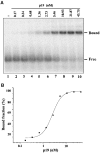
RNA silencing is an evolutionarily conserved surveillance system that occurs in a broad range of eukaryotic organisms. In plants, RNA silencing acts as an antiviral system; thus, successful virus infection requires suppression of gene silencing. A number of viral suppressors have been identified so far; however, the molecular bases of silencing suppression are still poorly understood. Here we show that p19 of Cymbidium ringspot virus (CymRSV) inhibits RNA silencing via its small RNA-binding activity in vivo. Small RNAs bound by p19 in planta are bona fide double-stranded siRNAs and they are silencing competent in the in vitro RNA-silencing system. p19 also suppresses RNA silencing in the heterologous Drosophila in vitro system by preventing siRNA incorporation into RISC. During CymRSV infection, p19 markedly diminishes the amount of free siRNA in cells by forming p19-siRNA complexes, thus making siRNAs inaccessible for effector complexes of RNA-silencing machinery. Furthermore, the obtained results also suggest that the p19-mediated sequestration of siRNAs in virus-infected cells blocks the spread of the mobile, systemic signal of RNA silencing.
Figures



Similar articles
-
Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity.J Virol. 2005 Jun;79(11):7217-26. doi: 10.1128/JVI.79.11.7217-7226.2005. J Virol. 2005. PMID: 15890960 Free PMC article.
-
Distinct Effects of p19 RNA Silencing Suppressor on Small RNA Mediated Pathways in Plants.PLoS Pathog. 2016 Oct 6;12(10):e1005935. doi: 10.1371/journal.ppat.1005935. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27711201 Free PMC article.
-
Viral RNAi suppressor reversibly binds siRNA to outcompete Dicer and RISC via multiple turnover.J Mol Biol. 2011 Apr 29;408(2):262-76. doi: 10.1016/j.jmb.2011.02.038. Epub 2011 Feb 24. J Mol Biol. 2011. PMID: 21354178 Free PMC article.
-
Studying the RNA silencing pathway with the p19 protein.FEBS Lett. 2013 Apr 17;587(8):1198-205. doi: 10.1016/j.febslet.2013.01.036. Epub 2013 Jan 29. FEBS Lett. 2013. PMID: 23376479 Review.
-
Plant RNAi: How a viral silencing suppressor inactivates siRNA.Curr Biol. 2004 Mar 9;14(5):R198-200. doi: 10.1016/j.cub.2004.02.021. Curr Biol. 2004. PMID: 15028237 Review.
Cited by
-
SlS5H silencing reveals specific pathogen-triggered salicylic acid metabolism in tomato.BMC Plant Biol. 2022 Nov 29;22(1):549. doi: 10.1186/s12870-022-03939-5. BMC Plant Biol. 2022. PMID: 36443652 Free PMC article.
-
A tomato bushy stunt virus-based vector for simultaneous editing and sensing to survey the host antiviral RNA silencing machinery.PNAS Nexus. 2023 Dec 14;3(1):pgad436. doi: 10.1093/pnasnexus/pgad436. eCollection 2024 Jan. PNAS Nexus. 2023. PMID: 38264147 Free PMC article.
-
AC5 protein encoded by squash leaf curl China virus is an RNA silencing suppressor and a virulence determinant.Front Microbiol. 2022 Aug 19;13:980147. doi: 10.3389/fmicb.2022.980147. eCollection 2022. Front Microbiol. 2022. PMID: 36060769 Free PMC article.
-
Non-coding RNAs in Intercellular and Systemic Signaling.Front Plant Sci. 2012 Jul 5;3:141. doi: 10.3389/fpls.2012.00141. eCollection 2012. Front Plant Sci. 2012. PMID: 22783264 Free PMC article. No abstract available.
-
Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein.J Virol. 2007 Oct;81(19):10379-88. doi: 10.1128/JVI.00727-07. Epub 2007 Jul 18. J Virol. 2007. PMID: 17634237 Free PMC article.
References
-
- Ahlquist P (2002) RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273 - PubMed
-
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 - PubMed
-
- Baulcombe D (2002) Viral suppression of systemic silencing. Trends Microbiol 10: 306–308 - PubMed
-
- Beclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

