Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation
- PMID: 14976550
- PMCID: PMC380967
- DOI: 10.1038/sj.emboj.7600102
Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation
Abstract
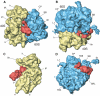
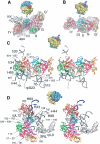
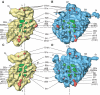
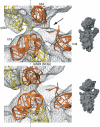
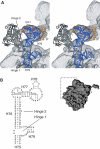
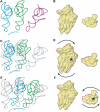
An 11.7-A-resolution cryo-EM map of the yeast 80S.eEF2 complex in the presence of the antibiotic sordarin was interpreted in molecular terms, revealing large conformational changes within eEF2 and the 80S ribosome, including a rearrangement of the functionally important ribosomal intersubunit bridges. Sordarin positions domain III of eEF2 so that it can interact with the sarcin-ricin loop of 25S rRNA and protein rpS23 (S12p). This particular conformation explains the inhibitory action of sordarin and suggests that eEF2 is stalled on the 80S ribosome in a conformation that has similarities with the GTPase activation state. A ratchet-like subunit rearrangement (RSR) occurs in the 80S.eEF2.sordarin complex that, in contrast to Escherichia coli 70S ribosomes, is also present in vacant 80S ribosomes. A model is suggested, according to which the RSR is part of a mechanism for moving the tRNAs during the translocation reaction.
Figures
References
-
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol 6: 643–647 - PubMed
-
- Ballesta JP, Remacha M (1996) The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog Nucleic Acid Res Mol Biol 55: 157–193 - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 - PubMed
-
- Beckmann R, Spahn CMT, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54 (Part 5): 905–921 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

