Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch
- PMID: 14976553
- PMCID: PMC380972
- DOI: 10.1038/sj.emboj.7600115
Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch
Abstract
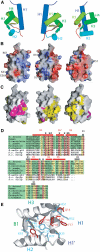
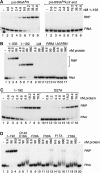
The La protein is a ubiquitous nuclear phosphoprotein that recognizes the 3' uridylates found in all newly synthesized RNA polymerase III transcripts. La binding stabilizes these transcripts from exonucleases and may also assist their folding. Here we present the first structural insights into how the La protein specifically interacts with its RNA substrates. The most conserved region of the La protein is the La motif, a domain also found in several other RNA-binding proteins. We have determined the structure of the La motif from the Trypanosoma brucei La protein to 1.6 A resolution (PDB code 1S29). The La motif adopts a winged helix-turn-helix architecture that has a highly conserved patch of mainly aromatic surface residues. Mutagenesis experiments support a critical role for this patch in RNA binding and show that it partly determines binding specificity for RNAs ending in 3' hydroxyl, a defining characteristic of the La protein. These findings reveal that the La motif is essential for high-affinity binding and also contributes to specificity.
Figures
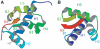

References
-
- Aigner S, Postberg J, Lipps HJ, Cech TR (2003) The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochem 42: 5736–5747 - PubMed
-
- Allers J, Shamoo Y (2001) Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J Mol Biol 311: 75–86 - PubMed
-
- Antson A (2000) Single stranded RNA binding proteins. Curr Opin Struct Biol 10: 87–94 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

