Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis
- PMID: 14977982
- PMCID: PMC356042
- DOI: 10.1128/IAI.72.3.1733-1745.2004
Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis
Abstract
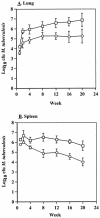
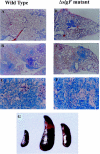
The Mycobacterium tuberculosis alternate sigma factor, SigF, is expressed during stationary growth phase and under stress conditions in vitro. To better understand the function of SigF we studied the phenotype of the M. tuberculosis DeltasigF mutant in vivo during mouse infection, tested the mutant as a vaccine in rabbits, and evaluated the mutant's microarray expression profile in comparison with the wild type. In mice the growth rates of the DeltasigF mutant and wild-type strains were nearly identical during the first 8 weeks after infection. At 8 weeks, the DeltasigF mutant persisted in the lung, while the wild type continued growing through 20 weeks. Histopathological analysis showed that both wild-type and mutant strains had similar degrees of interstitial and granulomatous inflammation during the first 12 weeks of infection. However, from 12 to 20 weeks the mutant strain showed smaller and fewer lesions and less inflammation in the lungs and spleen. Intradermal vaccination of rabbits with the M. tuberculosis DeltasigF strain, followed by aerosol challenge, resulted in fewer tubercles than did intradermal M. bovis BCG vaccination. Complete genomic microarray analysis revealed that 187 genes were relatively underexpressed in the absence of SigF in early stationary phase, 277 in late stationary phase, and only 38 genes in exponential growth phase. Numerous regulatory genes and those involved in cell envelope synthesis were down-regulated in the absence of SigF; moreover, the DeltasigF mutant strain lacked neutral red staining, suggesting a reduction in the expression of envelope-associated sulfolipids. Examination of 5'-untranslated sequences among the downregulated genes revealed multiple instances of a putative SigF consensus recognition sequence: GGTTTCX(18)GGGTAT. These results indicate that in the mouse the M. tuberculosis DeltasigF mutant strain persists in the lung but at lower bacterial burdens than wild type and is attenuated by histopathologic assessment. Microarray analysis has identified SigF-dependent genes and a putative SigF consensus recognition site.
Figures

Similar articles
-
Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF.Infect Immun. 2000 Oct;68(10):5575-80. doi: 10.1128/IAI.68.10.5575-5580.2000. Infect Immun. 2000. PMID: 10992456 Free PMC article.
-
The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence.Cell Microbiol. 2005 Feb;7(2):233-44. doi: 10.1111/j.1462-5822.2004.00454.x. Cell Microbiol. 2005. PMID: 15659067
-
Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network.J Bacteriol. 2008 Jan;190(2):699-707. doi: 10.1128/JB.01273-07. Epub 2007 Nov 9. J Bacteriol. 2008. PMID: 17993538 Free PMC article.
-
Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set.Mol Microbiol. 2004 Apr;52(1):25-38. doi: 10.1111/j.1365-2958.2003.03958.x. Mol Microbiol. 2004. PMID: 15049808
-
Characterization of the Mycobacterium tuberculosis sigma factor SigM by assessment of virulence and identification of SigM-dependent genes.Infect Immun. 2007 Jan;75(1):452-61. doi: 10.1128/IAI.01395-06. Epub 2006 Nov 6. Infect Immun. 2007. PMID: 17088352 Free PMC article.
Cited by
-
Mycobacterial Regulatory Systems Involved in the Regulation of Gene Expression Under Respiration-Inhibitory Conditions.J Microbiol. 2023 Mar;61(3):297-315. doi: 10.1007/s12275-023-00026-8. Epub 2023 Feb 27. J Microbiol. 2023. PMID: 36847970 Review.
-
The Partner Switching System of the SigF Sigma Factor in Mycobacterium smegmatis and Induction of the SigF Regulon Under Respiration-Inhibitory Conditions.Front Microbiol. 2020 Nov 11;11:588487. doi: 10.3389/fmicb.2020.588487. eCollection 2020. Front Microbiol. 2020. PMID: 33304334 Free PMC article.
-
The yeast-phase virulence requirement for α-glucan synthase differs among Histoplasma capsulatum chemotypes.Eukaryot Cell. 2011 Jan;10(1):87-97. doi: 10.1128/EC.00214-10. Epub 2010 Oct 29. Eukaryot Cell. 2011. PMID: 21037179 Free PMC article.
-
Mycobacterium tuberculosis Transcription Machinery: Ready To Respond to Host Attacks.J Bacteriol. 2016 Apr 14;198(9):1360-73. doi: 10.1128/JB.00935-15. Print 2016 May. J Bacteriol. 2016. PMID: 26883824 Free PMC article. Review.
-
Reconstruction and topological characterization of the sigma factor regulatory network of Mycobacterium tuberculosis.Nat Commun. 2016 Mar 31;7:11062. doi: 10.1038/ncomms11062. Nat Commun. 2016. PMID: 27029515 Free PMC article.
References
-
- Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control sigmaF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. - PubMed
-
- Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. - PubMed
-
- Belanger, A. E., and J. M. Inamine. 2000. Genetics of cell wall biosynthesis, p. 191-202. In G. E. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
-
- Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

