CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis
- PMID: 14978263
- PMCID: PMC365783
- DOI: 10.1073/pnas.0400163101
CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis
Erratum in
- Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):6326
Abstract
The circadian clock controls numerous physiological and molecular processes in organisms ranging from fungi to human. In plants, these processes include leaf movement, stomata opening, and expression of a large number of genes. At the core of the circadian clock, the central oscillator consists of a negative autoregulatory feedback loop that is coordinated with the daily environmental changes, and that generates the circadian rhythms of the overt processes. Phosphorylation of some of the central oscillator proteins is necessary for the generation of normal circadian rhythms of Drosophila, humans, and Neurospora, where CK1 and CK2 are emerging as the main protein kinases involved in the phosphorylation of PER and FRQ. We have previously shown that in Arabidopsis, the protein kinase CK2 can phosphorylate the clock-associated protein CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) in vitro. The overexpression of one of its regulatory subunits, CKB3, affects the regulation of circadian rhythms. Whether the effects of CK2 on the clock were due to its phosphorylation of a clock component had yet to be proven. By examining the effects of constitutively expressing a mutant form of the Arabidopsis clock protein CCA1 that cannot be phosphorylated by CK2, we demonstrate here that CCA1 phosphorylation by CK2 is important for the normal functioning of the central oscillator.
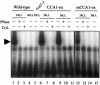
Figures



References
-
- Barak, S., Tobin, E. M., Andronis, C., Sugano, S. & Green, R. M. (2000) Trends Plant Sci. 5, 517-522. - PubMed
-
- Wang, Z. Y. & Tobin, E. M. (1998) Cell 93, 1207-1217. - PubMed
-
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I. A. & Coupland, G. (1998) Cell 93, 1219-1229. - PubMed
-
- Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P. & Kay, S. A. (2001) Science 293, 880-883. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

