Dimethyl sulfoxide at 2.5% (v/v) alters the structural cooperativity and unfolding mechanism of dimeric bacterial NAD+ synthetase
- PMID: 14978314
- PMCID: PMC2286739
- DOI: 10.1110/ps.03330104
Dimethyl sulfoxide at 2.5% (v/v) alters the structural cooperativity and unfolding mechanism of dimeric bacterial NAD+ synthetase
Abstract
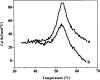
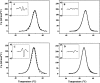
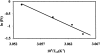
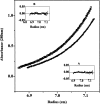
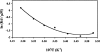
Dimethyl sulfoxide (DMSO) is commonly used as a cosolvent to improve the aqueous solubility of small organic compounds. Its use in a screen to identify novel inhibitors of the enzyme NAD(+) synthetase led to this investigation of its potential effects on the structure and stability of this 60-kD homodimeric enzyme. Although no effects are observed on the enzyme's catalytic activity, as low as 2.5% (v/v) DMSO led to demonstrable changes in the stability of the dimer and its unfolding mechanism. In the absence of DMSO, the dimer behaves hydrodynamically as a single ideal species, as determined by equilibrium analytical ultracentrifugation, and thermally unfolds according to a two-state dissociative mechanism, based on analysis by differential scanning calorimetry (DSC). In the presence of 2.5% (v/v) DMSO, an equilibrium between the dimer and monomer is now detectable with a measured dimer association constant, K(a), equal to 5.6 x 10(6)/M. DSC curve analysis is consistent with this finding. The data are best fit to a three-state sequential unfolding mechanism, most likely representing folded dimer <==> folded monomer <==> unfolded monomer. The unusually large change in the relative stabilities of dimer and monomer, e.g., the association equilibrium shifts from an infinitely large K(a) down to approximately 10(6)/M, in the presence of relatively low cosolvent concentration is surprising in view of the significant buried surface area at the dimer interface, roughly 20% of the surface area of each monomer is buried. A hypothetical structural mechanism to explain this effect is presented.
Figures


Similar articles
-
Thermodynamic analysis of unfolding and dissociation in lactose repressor protein.Biochemistry. 1999 May 18;38(20):6520-8. doi: 10.1021/bi9900727. Biochemistry. 1999. PMID: 10350470
-
Equilibrium unfolding of dimeric and engineered monomeric forms of lambda Cro (F58W) repressor and the effect of added salts: evidence for the formation of folded monomer induced by sodium perchlorate.Arch Biochem Biophys. 2005 Feb 1;434(1):93-107. doi: 10.1016/j.abb.2004.10.019. Arch Biochem Biophys. 2005. PMID: 15629113
-
Detection of early unfolding events in a dimeric protein by amide proton exchange and native electrospray mass spectrometry.Protein Sci. 2009 Aug;18(8):1620-7. doi: 10.1002/pro.176. Protein Sci. 2009. PMID: 19554628 Free PMC article.
-
Hyperthermophile protein folding thermodynamics: differential scanning calorimetry and chemical denaturation of Sac7d.J Mol Biol. 1996 Dec 13;264(4):784-805. doi: 10.1006/jmbi.1996.0677. J Mol Biol. 1996. PMID: 8980686
-
DMSO-Induced Unfolding of the Antifungal Disulfide Protein PAF and Its Inactive Variant: A Combined NMR and DSC Study.Int J Mol Sci. 2023 Jan 7;24(2):1208. doi: 10.3390/ijms24021208. Int J Mol Sci. 2023. PMID: 36674720 Free PMC article.
Cited by
-
Stability of the Retinoid X Receptor-α Homodimer in the Presence and Absence of Rexinoid and Coactivator Peptide.Biochemistry. 2021 Apr 20;60(15):1165-1177. doi: 10.1021/acs.biochem.0c00865. Epub 2021 Apr 1. Biochemistry. 2021. PMID: 33792309 Free PMC article.
-
Structural features differentiate the mechanisms between 2S (2 state) and 3S (3 state) folding homodimers.Bioinformation. 2005 Sep 2;1(2):42-9. doi: 10.6026/97320630001042. Bioinformation. 2005. PMID: 17597851 Free PMC article.
-
The calcium-induced conformation and glycosylation of scavenger-rich cysteine repeat (SRCR) domains of glycoprotein 340 influence the high affinity interaction with antigen I/II homologs.J Biol Chem. 2014 Aug 8;289(32):21877-87. doi: 10.1074/jbc.M114.565507. Epub 2014 Jun 12. J Biol Chem. 2014. PMID: 24923446 Free PMC article.
-
Coregulation of Terpenoid Pathway Genes and Prediction of Isoprene Production in Bacillus subtilis Using Transcriptomics.PLoS One. 2013 Jun 19;8(6):e66104. doi: 10.1371/journal.pone.0066104. Print 2013. PLoS One. 2013. PMID: 23840410 Free PMC article.
-
Structural and biophysical characterization of the α-carbonic anhydrase from the gammaproteobacterium Thiomicrospira crunogena XCL-2: insights into engineering thermostable enzymes for CO2 sequestration.Acta Crystallogr D Biol Crystallogr. 2015 Aug;71(Pt 8):1745-56. doi: 10.1107/S1399004715012183. Epub 2015 Jul 31. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 26249355 Free PMC article.
References
-
- Azuaga, A.I., Conejero-Lara, F., Rivas, G., De Filippis, V., Fontana, A., and Mateo, P.L. 1995. The thermodynamics of association and unfolding of the 205–316 C-terminal fragment of thermolysin. Biochim. Biophys. Acta 1252 95–102. - PubMed
-
- Baker, B.M. and Murphy, K.P. 1998. Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 295 294–315. - PubMed
-
- Bowie, J.U. and Sauer, R.T. 1989. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry 28 7139–7143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

