Novel approaches to gene expression analysis of active polyarticular juvenile rheumatoid arthritis
- PMID: 14979934
- PMCID: PMC400410
- DOI: 10.1186/ar1018
Novel approaches to gene expression analysis of active polyarticular juvenile rheumatoid arthritis
Abstract
Juvenile rheumatoid arthritis (JRA) has a complex, poorly characterized pathophysiology. Modeling of transcriptosome behavior in pathologic specimens using microarrays allows molecular dissection of complex autoimmune diseases. However, conventional analyses rely on identifying statistically significant differences in gene expression distributions between patients and controls. Since the principal aspects of disease pathophysiology vary significantly among patients, these analyses are biased. Genes with highly variable expression, those most likely to regulate and affect pathologic processes, are excluded from selection, as their distribution among healthy and affected individuals may overlap significantly. Here we describe a novel method for analyzing microarray data that assesses statistically significant changes in gene behavior at the population level. This method was applied to expression profiles of peripheral blood leukocytes from a group of children with polyarticular JRA and healthy control subjects. Results from this method are compared with those from a conventional analysis of differential gene expression and shown to identify discrete subsets of functionally related genes relevant to disease pathophysiology. These results reveal the complex action of the innate and adaptive immune responses in patients and specifically underscore the role of IFN-gamma in disease pathophysiology. Discriminant function analysis of data from a cohort of patients treated with conventional therapy identified additional subsets of functionally related genes; the results may predict treatment outcomes. While data from only 9 patients and 12 healthy controls was used, this preliminary investigation of the inflammatory genomics of JRA illustrates the significant potential of utilizing complementary sets of bioinformatics tools to maximize the clinical relevance of microarray data from patients with autoimmune disease, even in small cohorts.
Figures



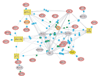
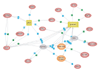


References
-
- Jarvis JN. Pathogenesis and mechanisms of inflammation in the childhood rheumatic diseases. Curr Opin Rheumatol. 1998;10:459–467. - PubMed
-
- Lo D, Feng L, Li L, Carson MJ, Crowley M, Pauza M, Nguyen A, Reilly CR. Integrating innate and adaptive immunity in the whole animal. Immunol Rev. 1999;169:225–239. - PubMed
-
- Fearon DT, Locksley RM. The instructive role of innate immunity in the acquired immune response. Science. 1996;72:50–53. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

