Determinants of liposome fusion mediated by synaptic SNARE proteins
- PMID: 14981239
- PMCID: PMC365710
- DOI: 10.1073/pnas.0400044101
Determinants of liposome fusion mediated by synaptic SNARE proteins
Abstract
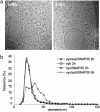
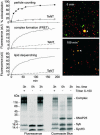
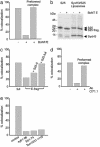
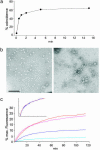
Synaptic exocytosis requires the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins syntaxin 1, SNAP-25, and synaptobrevin (VAMP). Assembly of the SNAREs into a stable core complex is supposed to catalyze membrane fusion, and proteoliposomes reconstituted with synaptic SNARE proteins spontaneously fuse with each other. We now show that liposome fusion mediated by synaptic SNAREs is inhibited by botulinum neurotoxin E (BoNT/E) but can be rescued by supplementing the C-terminal portion of SNAP-25. Furthermore, fusion is prevented by a SNAP-25-specific antibody known to block exocytosis in chromaffin cells, and it is competed for by soluble fragments of the R-SNAREs synaptobrevin 2, endobrevin/VAMP-8, and tomosyn. No accumulation of clustered vesicles is observed during the reaction. Rapid artificial clustering of SNARE-containing proteoliposomes enhances the fusion rate at low but not at saturating liposome concentrations. We conclude that the rate of liposome fusion is dominated by the intrinsic properties of the SNAREs rather than by the preceding docking step.
Figures

References
-
- Chen, Y. A. & Scheller, R. H. (2001) Nat. Rev. Mol. Cell. Biol. 2, 98-106. - PubMed
-
- Jahn, R., Lang, T. & Sudhof, T. C. (2003) Cell 112, 519-533. - PubMed
-
- Rizo, J. & Sudhof, T. C. (2002) Nat. Rev. Neurosci. 3, 641-653. - PubMed
-
- Sutton, B., Fasshauer, D., Jahn, R. & Brünger, A. T. (1998) Nature 395, 347-353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

