The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes
- PMID: 14982958
- PMCID: PMC390273
- DOI: 10.1093/nar/gkh277
The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes
Abstract
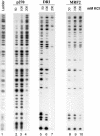
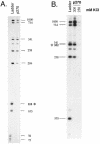
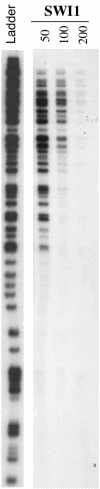
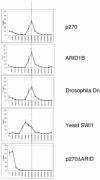
SWI/SNF complexes are ATP-dependent chromatin remodeling complexes that are highly conserved from yeast to human. From yeast to human the complexes contain a subunit with an ARID (A-T-rich interaction domain) DNA-binding domain. In yeast this subunit is SWI1 and in human there are two closely related alternative subunits, p270 and ARID1B. We describe here a comparison of the DNA-binding properties of the yeast and human SWI/SNF ARID-containing subunits. We have determined that SWI1 is an unusual member of the ARID family in both its ARID sequence and in the fact that its DNA-binding affinity is weaker than that of other ARID family members, including its human counterparts, p270 and ARID1B. Sequence analysis and substitution mutagenesis reveals that the weak DNA-binding affinity of the SWI1 ARID is an intrinsic feature of its sequence, arising from specific variations in the major groove interaction site. In addition, this work confirms the finding that p270 binds DNA without regard to sequence specificity, excluding the possibility that the intrinsic role of the ARID is to recruit SWI/SNF complexes to specific promoter sequences. These results emphasize that care must be taken when comparing yeast and higher eukaryotic SWI/SNF complexes in terms of DNA-binding mechanisms.
Figures



References
-
- Martens J.A. and Winston,F. (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev., 13, 136–142. - PubMed
-
- Quinn J., Fyrberg,A.M., Ganster,R.W., Schmidt,M.C. and Peterson,C.L. (1996) DNA-binding properties of the yeast SWI/SNF complex. Nature, 379, 844–847. - PubMed
-
- Sengupta S.M., Van Kanegan,M., Persinger,J., Logie,C., Cairns,B.R., Peterson,C.L. and Bartholomew,B. (2001) The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem., 276, 12636–12644. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

