Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression
- PMID: 14983020
- PMCID: PMC356961
- DOI: 10.1073/pnas.0308703100
Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression
Abstract
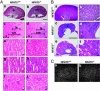
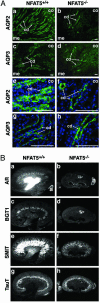
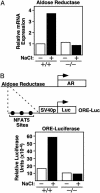
The transcription factor NFAT5/TonEBP, a member of the NFAT/Rel family of transcription factors, has been implicated in diverse cellular responses, including the response to osmotic stress, integrin-dependent cell migration, T cell activation, and the Ras pathway in Drosophila. To clarify the in vivo role of NFAT5, we generated NFAT5-null mice. Homozygous mutants were genetically underrepresented after embryonic day 14.5. Surviving mice manifested a progressive and profound atrophy of the kidney medulla with impaired activation of several osmoprotective genes, including those encoding aldose reductase, Na+/Cl--coupled betaine/gamma-aminobutyric acid transporter, and the Na+/myo-inositol cotransporter. The aldose reductase gene is controlled by a tonicity-responsive enhancer, which was refractory to hypertonic stress in fibroblasts lacking NFAT5, establishing this enhancer as a direct transcriptional target of NFAT5. Our findings demonstrate a central role for NFAT5 as a tonicity-responsive transcription factor required for kidney homeostasis and function.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

