Evolutionary comparisons suggest many novel cAMP response protein binding sites in Escherichia coli
- PMID: 14983022
- PMCID: PMC356963
- DOI: 10.1073/pnas.0308628100
Evolutionary comparisons suggest many novel cAMP response protein binding sites in Escherichia coli
Abstract
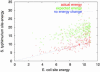
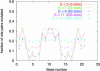
The cAMP response protein (CRP) is a transcription factor known to regulate many genes in Escherichia coli. Computational studies of transcription factor binding to DNA are usually based on a simple matrix model of sequence-dependent binding energy. For CRP, this model predicts many binding sites that are not known to be functional. If they are indeed spurious, the underlying binding model is called into question. We use a species comparison method to assess the functionality of a population of such predicted CRP sites in E. coli. We compare them with orthologous sites in Salmonella typhimurium identified independently by CLUSTALW alignment, and find a dependence of mutation probability on position in the site. This dependence increases with predicted site binding energy. The positions where mutation is most strongly suppressed are those where mutation would have the biggest effect on predicted binding energy. This finding suggests that many of the novel sites are functional, that the matrix model correctly estimates their binding strength, and that calculated CRP binding strength is the quantity that is conserved between species. The analysis also identifies many new E. coli binding sites and genes likely to be functional for CRP.
Figures
References
-
- Ptashne, M. (1992) A Genetic Switch (Blackwell Scientific, Oxford).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous