Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake
- PMID: 14988478
- PMCID: PMC389937
- DOI: 10.1104/pp.103.034660
Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake
Abstract
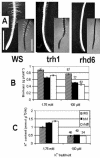
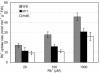
Potassium (K(+)) is the most abundant cation in plants and is required for plant growth. To ensure an adequate supply of K(+), plants have multiple mechanisms for uptake and translocation. However, relatively little is known about the physiological role of proteins encoded by a family of 13 genes, named AtKT/KUP, that are involved in K(+) transport and translocation. To begin to understand where and under what conditions these transporters function, we used reverse transcription-PCR to determine the spatial and temporal expression patterns of each AtKT/KUP gene across a range of organs and tested whether selected AtKT/KUP cDNAs function as K(+) transporters in Escherichia coli. Many AtKT/KUPs were expressed in roots, leaves, siliques, and flowers of plants grown under K(+)-sufficient conditions (1.75 mm KCl) in hydroponic culture. AtHAK5 was the only gene in this family that was up-regulated upon K(+) deprivation and rapidly down-regulated with resupply of K(+). Ten AtKT/KUPs were expressed in root hairs, but only five were expressed in root tip cells. This suggests an important role for root hairs in K(+) uptake. The growth and rubidium (Rb(+)) uptake of two root hair mutants, trh1-1 (tiny root hairs) and rhd6 (root hair defective), were studied to determine the contribution of root hairs to whole-plant K(+) status. Whole-plant biomass decreased in the root hair mutants only when K(+) concentrations were low; Rb(+) (used as a tracer for K(+)) uptake rates were lower in the mutants at all Rb(+) concentrations. Seven genes encoding AtKUP transporters were expressed in E. coli (AtKT3/KUP4, AtKT/KUP5, AtKT/KUP6, AtKT/KUP7, AtKT/KUP10, AtKT/KUP11, and AtHAK5), and their K(+) transport function was demonstrated.
Figures





References
-
- Bates TR, Lynch JP (2000) Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). Am J Bot 87: 958-963 - PubMed
-
- Buurman ET, Kim KT, Epstein W (1995) Genetic evidence for two sequentially occupied K+ binding sites in the Kdp transport ATPase. J Biol Chem 270: 6678-6685 - PubMed
-
- Deeken R, Geiger D, Fromm J, Koroleva O, Ache P, Langenfeld-Heyser R, Sauer N, May ST, Hedrich R (2002) Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216: 334-344 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

