Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins
- PMID: 14988731
- PMCID: PMC381411
- DOI: 10.1038/sj.emboj.7600137
Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins
Abstract
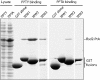
Membrane proteins destined for the vacuolar or lysosomal lumen are typically ubiquitinated, the ubiquitin serving as a targeting signal for the multivesicular body pathway. The RING-domain ubiquitin ligase Tul1 is an integral membrane protein that modifies the yeast vacuolar enzyme carboxypeptidase S (Cps1), the polyphosphatase Ppn1/Phm5 and other proteins containing exposed hydrophilic residues within their transmembrane domains (TMDs). Here we show that Bsd2 provides an alternative ubiquitination mechanism for Cps1, Phm5 and other proteins. Bsd2 is a three-TMD protein with a PPXY motif that binds the HECT domain ubiquitin ligase Rsp5. It can thus act as a specific adaptor linking Rsp5 to its substrates. Like Tul1, the Bsd2 system recognises polar TMDs. Bsd2 also controls the vacuolar targeting of a manganese transporter and a mutant plasma membrane ATPase, and together with the ER retrieval receptor Rer1, it protects cells from stress. We suggest that Bsd2 has a wide role in the quality control of membrane proteins. Bsd2 is the yeast homologue of human NEDD4 binding protein N4WBP5, which may therefore have similar functions.
Figures






References
-
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC (2002) The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol 4: 534–539 - PubMed
-
- Cristillo AD, Nie L, Macri MJ, Bierer BE (2003) Cloning and characterization of N4WBP5A, an inducible, cyclosporine-sensitive, Nedd4-binding protein in human T lymphocytes. J Biol Chem 278: 34587–34597 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

