A dual phenotype of periventricular nodular heterotopia and frontometaphyseal dysplasia in one patient caused by a single FLNA mutation leading to two functionally different aberrant transcripts
- PMID: 14988809
- PMCID: PMC1181949
- DOI: 10.1086/383094
A dual phenotype of periventricular nodular heterotopia and frontometaphyseal dysplasia in one patient caused by a single FLNA mutation leading to two functionally different aberrant transcripts
Abstract
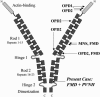
Two disorders, periventricular nodular heterotopia (PVNH) and a group of skeletal dysplasias belonging to the oto-palato-digital (OPD) spectrum, are caused by FLNA mutations. They are considered mutually exclusive because of the different presumed effects of the respective FLNA gene mutations, leading to loss of function (PVNH) and gain of function (OPD), respectively. We describe here the first patient manifesting PVNH in combination with frontometaphyseal dysplasia, a skeletal dysplasia of the OPD-spectrum. A novel de novo mutation, 7315C-->A in exon 45 of the FLNA gene, was identified. It leads to two aberrant transcripts, one full-length transcript with the point mutation causing a substitution of a highly conserved leucine residue (L2439M) and a second shortened transcript lacking 21 bp due to the creation of an ectopic splice donor site in exon 45. We propose that the dual phenotype is caused by two functionally different, aberrant filamin A proteins and therefore represents an exceptional model case of allelic gain-of-function and loss-of-function phenotypes due to a single mutational event.
Figures




References
Electronic-Database Information
-
- dbSNP Home Page, http://www.ncbi.nih.gov/SNP/ (for IVS45-64T→C [accession number rs2070819] and coding SNPs within FLNA [accession number rs2070825] and TIMP1 [accession number rs4898])
-
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for FLNA [accession numbers: genomic: NT_025965; mRNA: NM_001456; protein: NP_001447; GenBank: X53416], FLNA protein [Mus musculus: gi|13278531|gb|AAH04061.1], filamin A [Bos taurus: gi|18377580|gb|AAL66773.1], for filamin A [Gallus gallus: gi|15341202|dbj|BAB63943.1], filamin C [Homo sapiens: gi|4218955|gb|AAD12245.1], filamin B [H. sapiens: gi|3298597|gb|AAC39842.1], filamin1 [Drosophila melanogaster: gi|6707288|gb|AAF25614.1|AF174492_1], Filamin/ABP280 repeat family member (4B112) [Caenorhabditis elegans: gi|17543918|ref|NP_499911.1])
-
- Human Gene Mutation Database, http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html (for known FLNA mutations in PVNH)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PVNH, OPD1, OPD2, MNS, and FMD)
-
- Protein Data Bank, http://www.rcsb.org/pdb/ (for the structures of the 4th [1KSR] and 5th [1QFH] module of the F-actin cross-linking gelation factor [Abp-120])
References
-
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, Berkovic SF, Huttenlocher PR, Walsh CA (1998) Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21:1315–1325 - PubMed
-
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA (1997) The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat Struct Biol 4:223–230 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

