Insights into the molecular mechanism of rotation in the Fo sector of ATP synthase
- PMID: 14990464
- PMCID: PMC1303972
- DOI: 10.1016/S0006-3495(04)74205-8
Insights into the molecular mechanism of rotation in the Fo sector of ATP synthase
Abstract
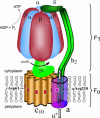
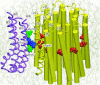
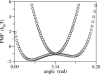
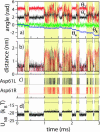
F(1)F(o)-ATP synthase is a ubiquitous membrane protein complex that efficiently converts a cell's transmembrane proton gradient into chemical energy stored as ATP. The protein is made of two molecular motors, F(o) and F(1), which are coupled by a central stalk. The membrane unit, F(o), converts the transmembrane electrochemical potential into mechanical rotation of a rotor in F(o) and the physically connected central stalk. Based on available data of individual components, we have built an all-atom model of F(o) and investigated through molecular dynamics simulations and mathematical modeling the mechanism of torque generation in F(o). The mechanism that emerged generates the torque at the interface of the a- and c-subunits of F(o) through side groups aSer-206, aArg-210, and aAsn-214 of the a-subunit and side groups cAsp-61 of the c-subunits. The mechanism couples protonation/deprotonation of two cAsp-61 side groups, juxtaposed to the a-subunit at any moment in time, to rotations of individual c-subunit helices as well as rotation of the entire c-subunit. The aArg-210 side group orients the cAsp-61 side groups and, thereby, establishes proton transfer via aSer-206 and aAsn-214 to proton half-channels, while preventing direct proton transfer between the half-channels. A mathematical model proves the feasibility of torque generation by the stated mechanism against loads typical during ATP synthesis; the essential model characteristics, e.g., helix and subunit rotation and associated friction constants, have been tested and furnished by steered molecular dynamics simulations.
Figures






References
-
- Abrahams, J., A. Leslie, R. Lutter, and J. Walker. 1994. Structure at 2.8-Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 370:621–628. - PubMed
-
- Angevine, C. M., and R. H. Fillingame. 2003. Aqueous access channels in subunit a of rotary ATP synthase. J. Biol. Chem. 278:6066–6074. - PubMed
-
- Batcho, P. F., D. A. Case, and T. Schlick. 2001. Optimized particle-mesh Ewald/multiple-time step integration for molecular dynamics simulations. J. Chem. Phys. 115:4003–4018.
-
- Berg, H. editor. 1983. Random Walks in Biology. Princeton University Press, Princeton, NJ.
-
- Böckmann, R. A., and H. Grubmüller. 2002. Nanoseconds molecular dynamics simulation of primary mechanical energy transfer steps in F1-ATP synthase. Nat. Struct. Biol. 9:198–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

