Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor
- PMID: 14990699
- PMCID: PMC353740
- DOI: 10.1128/jvi.78.6.2790-2807.2004
Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor
Erratum in
- J Virol. 2005 Jun;78(12):6706
Abstract
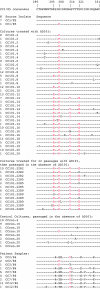
We have described previously the generation of an escape variant of human immunodeficiency virus type 1 (HIV-1), under the selection pressure of AD101, a small molecule inhibitor that binds the CCR5 coreceptor (A. Trkola, S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. X. L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore, Proc. Natl. Acad. Sci. USA 99:395-400, 2002). The escape mutant, CC101.19, continued to use CCR5 for entry, but it was at least 20,000-fold more resistant to AD101 than the parental virus, CC1/85. We have now cloned the env genes from the the parental and escape mutant isolates and made chimeric infectious molecular clones that fully recapitulate the phenotypes of the corresponding isolates. Sequence analysis of the evolution of the escape mutants suggested that the most relevant changes were likely to be in the V3 loop of the gp120 glycoprotein. We therefore made a series of mutant viruses and found that full AD101 resistance was conferred by four amino acid changes in V3. Each change individually caused partial resistance when they were introduced into the V3 loop of a CC1/85 clone, but their impact was dependent on the gp120 context in which they were made. We assume that these amino acid changes alter how the HIV-1 Env complex interacts with CCR5. Perhaps unexpectedly, given the complete dependence of the escape mutant on CCR5 for entry, monomeric gp120 proteins expressed from clones of the fully resistant isolate failed to bind to CCR5 on the surface of L1.2-CCR5 cells under conditions where gp120 proteins from the parental virus and a partially AD101-resistant virus bound strongly. Hence, the full impact of the V3 substitutions may only be apparent at the level of the native Env complex.
Figures





References
-
- Aarons, E. J., S. Beddows, T. Willingham, L. Wu, and R. A. Koup. 2001. Adaptation to blockade of human immunodeficiency virus type 1 entry imposed by the anti-CCR5 monoclonal antibody 2D7. Virology 287:382-390. - PubMed
-
- Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M. C. Gauduin, R. A. Koup, J. S. McDougal, and P. J. Maddon. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 11:533-539. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Alves, K., M. Canzian, and E. L. Delwart. 2002. HIV type 1 envelope quasispecies in the thymus and lymph Nodes of AIDS patients. AIDS Res. Hum. Retrovir. 18:161-165. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

