Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution
- PMID: 14990750
- PMCID: PMC390300
- DOI: 10.1093/nar/gkh315
Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution
Abstract
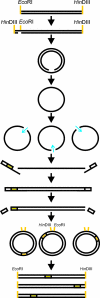
Directed molecular evolution and combinatorial methodologies are playing an increasingly important role in the field of protein engineering. The general approach of generating a library of partially randomized genes, expressing the gene library to generate the proteins the library encodes and then screening the proteins for improved or modified characteristics has successfully been applied in the areas of protein-ligand binding, improving protein stability and modifying enzyme selectivity. A wide range of techniques are now available for generating gene libraries with different characteristics. This review will discuss these different methodologies, their accessibility and applicability to non-expert laboratories and the characteristics of the libraries they produce. The aim is to provide an up to date resource to allow groups interested in using directed evolution to identify the most appropriate methods for their purposes and to guide those moving on from initial experiments to more ambitious targets in the selection of library construction techniques. References are provided to original methodology papers and other recent examples from the primary literature that provide details of experimental methods.
Figures



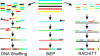
References
-
- Farinas E.T., Bulter,T. and Arnold,F.H. (2001) Directed enzyme evolution. Curr. Opin. Biotechnol., 12, 545–551. - PubMed
-
- Reetz M.T., Rentzsch,M., Pletsch,A. and Maywald,M. (2002) Towards the directed evolution of hybrid catalysts. Chimia, 56, 721–723.
-
- Taylor S.V., Kast,P. and Hilvert,D. (2001) Investigating and engineering enzymes by genetic selection. Angew. Chem. Int. Ed. Engl., 40, 3310–3335. - PubMed
-
- Waldo G.S. (2003) Genetic screens and directed evolution for protein solubility. Curr. Opin. Chem. Biol., 7, 33–38. - PubMed
-
- Tao H. and Cornish,V.W. (2002) Milestones in directed enzyme evolution. Curr. Opin. Chem. Biol., 6, 858–864. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

